-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK Pathway
In fungal species, differentiation to the filamentous/hyphal cell type is critical for entry into host cells and virulence. Comparative RNA sequencing was used to explore the pathways that regulate differentiation to the filamentous cell type in yeast. This approach uncovered a role for the stress-response MAPK pathway, HOG, during the increased metabolic respiration that induces filamentous growth. In this context, the AMPK Snf1p and ER stress kinase Ire1p regulated the HOG pathway. Cross-modulation between the HOG and filamentous growth (ERK-type) MAPK pathways optimized the differentiation response. The regulatory circuit described here may extend to behaviors in metazoans.
Published in the journal: Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK Pathway. PLoS Genet 10(10): e32767. doi:10.1371/journal.pgen.1004734
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004734Summary
In fungal species, differentiation to the filamentous/hyphal cell type is critical for entry into host cells and virulence. Comparative RNA sequencing was used to explore the pathways that regulate differentiation to the filamentous cell type in yeast. This approach uncovered a role for the stress-response MAPK pathway, HOG, during the increased metabolic respiration that induces filamentous growth. In this context, the AMPK Snf1p and ER stress kinase Ire1p regulated the HOG pathway. Cross-modulation between the HOG and filamentous growth (ERK-type) MAPK pathways optimized the differentiation response. The regulatory circuit described here may extend to behaviors in metazoans.
Introduction
Organisms sense and respond to diverse stimuli through the action of signal transduction pathways. During complex behaviors like cell differentiation, multiple pathways choreograph changes in the cell cycle and cell polarity to produce a new cell type. In many cases, it is not clear what pathways are involved or how different pathways collaborate to produce major changes in cellular reprogramming. Here, we investigate the problem of cell differentiation in fungal species that differentiate to the filamentous/hyphal cell type. In pathogens, filamentous growth is critical for virulence [1], [2]. Therefore, identifying the pathways that regulate filamentous growth, and understanding how they function in an interconnected manner, is important for studies in eukaryotic cell differentiation and fungal pathogenesis.
In the budding yeast Saccharomyces cerevisiae, different MAPK pathways mediate the response to different stimuli. An ERK-type MAPK pathway called the filamentous growth pathway induces differentiation to the filamentous/invasive/pseudohyphal cell type in response to nutrient limitation [3], [4]. A p38-type MAPK pathway, called the high osmolarity glycerol response (HOG) pathway mediates osmoadaptation [5], [6]. The two pathways utilize some of the same components, including a core module consisting of the Rho-type GTPase Cdc42p, the p21 activated (PAK) kinase Ste20p, the MAPKKK Ste11p, and the adaptor protein Ste50p (Fig. S1, [5], [7]–[9] and references therein). Plasma membrane (PM) regulators of the filamentous growth pathway, Msb2p, Sho1p, and Opy2p [10]–[12], also regulate the Ste11p branch of the HOG pathway [13]–[15]. The signaling mucin Msb2p may preferentially regulate the filamentous growth pathway, as another signaling mucin, Hkr1p, has been shown to mainly regulate the HOG pathway [16], [17]. A second branch of the HOG pathway (Sln1p branch) converges on the MAPKK Pbs2p and is regulated by the two-component sensors Sln1p and Ypd1p, the protein kinase Ssk1p, and the redundant MAPKKKs Ssk2p and Ssk22p (Fig. S1, [13], [18]–[21]). Thus, different MAPK pathways mediate different responses through the action of common or shared signaling modules.
To date, the filamentous growth and HOG pathways are thought to control different responses [22]–[25]. The filamentous growth pathway induces differentiation into chains of branched interconnected filaments by regulating changes in the cell cycle [26]–[28], cell adhesion [29], [30], and cell polarity [31]–[33]. By comparison, the HOG pathway induces transient growth arrest [6], [34], [35] by the phosphorylation of translation initiation factors [36]–[38]. The HOG pathway also controls the production of osmolyte ([39] and references therein) and regulates changes in chromatin architecture [40]–[43], yet does not trigger a morphogenetic response. In fact, osmotic stress transiently depolarizes the actin cytoskeleton [44]–[46]. Further evidence that the pathways operate separate programs comes from the fact that the HOG pathway shuts off the filamentous growth pathway in response to osmotic shock [16], [47]–[49]. The filamentous growth pathway can likewise attenuate the HOG pathway [10].
The paradigm that signaling pathways (even ones that share components) function as separate entities is complicated by the fact that under certain conditions the concerted action of multiple MAPK pathways occurs. The HOG and protein kinase C (PKC, [50]) pathways are together required in some settings [51], where they act cooperatively [52] or sequentially [53]. During mating [54], [55], the PKC pathway maintains cell integrity [56], [57], and the HOG pathway balances changes in osmolarity [35]. Severe stress, like enzymatic digestion of the cell wall [58], [59] or defects in protein glycosylation [60] trigger multiple pathways as part of an ill-defined response. Diverse stimuli also trigger a general stress response, called the environmental stress response (ESR, [61], [62]) but how this is related to MAPK-dependent responses is not clear.
Here, comparative RNA sequencing (RNA seq) analysis was used to examine the transcriptional response to inducers of the HOG (salt) and filamentous growth (galactose) pathways. The response to glycosylation deficiency was also examined, because that stress requires the action of both pathways. Analysis of expression profiling data uncovered galactose as an inducer of the HOG pathway. Accordingly, the AMPK Snf1p and an intact TCA cycle were required to activate the HOG pathway during growth on galactose. The HOG pathway has recently been shown to be activated in response to ER stress and require the unfolded protein response (UPR) regulator Ire1p [63], [64]. Growth of cells in galactose induced Ire1p-dependent activation of the HOG pathway. Therefore, the HOG and filamentous growth pathways are both induced during growth on galactose. Both pathways contributed to growth under this condition, and remarkably, both pathways attenuated each other's activities. Modulation of the filamentous growth pathway by the HOG pathway was required to optimize the differentiation response. The regulatory circuit described here connects general regulators of cellular responses – AMPK, Ire1p, and MAPK (p38 and ERK) – in a regulatory axis that controls cell differentiation. This axis was conserved in other fungal species and may underlie differentiation-type responses in metazoans, which contain evolutionarily conserved regulatory pathways.
Results
Expression Profiling by RNA Seq Uncovers a Role for the HOG Pathway during Growth in Galactose
Comparative RNA sequencing (RNA seq, [65]) was performed to examine the response of S. cerevisiae cells to different stimuli. The response to osmotic stress (YEPD+0.4M KCl [23]), the non-preferred carbon source galactose (YEP-GAL, 2% GAL [66]), and an inhibitor of N-linked glycosylation (YEPD+2.5 µg tunicamycin [67]) were examined. Each stimulus induced the expression of overlapping and non-overlapping genes (Fig. 1A). As reported, salt induced targets of the HOG pathway (Table S1, [12], [24]), galactose induced the GAL genes and other starvation-responsive genes (Table S1, [68], [69]), and tunicamycin induced targets of the UPR and other genes (Table S1, [70], [71]). The different stimuli also induced an overlapping gene set (Fig. 1A, 504 genes). Common genes included targets of the ESR (150 of 504 total, Table S1 [61], [62]) and targets of the HOG pathway. Likewise, a partially overlapping set of repressed genes was also identified that included ESR targets (Fig. S2A).
Fig. 1. Gene expression profiling by RNA seq analysis and qPCR. 
A) Genes induced by salt, tunicamycin (TUN), or galactose (GAL). All RNA seq comparisons are provided in Table S1. B) Genes induced in a Pbs2p-dependent manner under the indicated conditions. Genes outlined by the dark blue circle (Pbs2p-dependent GAL specific) were functionally annotated in a pie chart in Fig. S2. C) Heat map of genes induced by the indicated stresses. Common targets and targets unique to each stimulus is shown. Asterisk, target of ESR. D) qPCR of HOG pathway target mRNAs in wild type and the pbs2Δ mutant grown in glucose (GLU, YEPD) and galactose (GAL, YEP-GAL). Error bars indicate +/−S.E.M. of three independent experiments. Actin (ACT1) mRNA was used as a control. E) Activity of p8XCRE-lacZ in wild-type cells (PC313) and pbs2Δ mutant (PC5035) grown in YEPD (5.5 hr), YEP-GAL (5.5 hr), and YEPD+0.4 M KCl (30 min). F) qPCR of Ste12p target mRNAs in wild type (PC538) and the ste12Δ (PC2382) mutant grown in glucose (YEPD) and galactose (YEP-GAL). See panel D for details. To explore the role of the HOG pathway in response to different stresses, RNA seq profiles were compared between wild-type cells and cells lacking the HOG pathway MAPKK Pbs2p [pbs2Δ]. An overlapping set of Pbs2p-dependent genes was induced in response to all three conditions (32 genes, Fig. 1, B and C). Many of the genes (Fig. 1C, 27/32, asterisks) are targets of the ESR. The HOG response to the different stimuli was complex: Pbs2p-dependent targets showed different levels of induction by the different inducers and different induction profiles. For example, different subsets of Pbs2p-dependent targets were unique to each stimulus (Fig. 1, B and C; Table S1) or common to two of the three stimuli (Fig. 1B). Thus, the HOG pathway actuates common and unique outputs in response to different stimuli.
The finding that galactose induced HOG pathway targets in a Pbs2p-dependent manner was unexpected. Pbs2p-dependent targets of the HOG pathway that were induced during growth in galactose regulate mitochondria/respiration, carbohydrate metabolism (including gluconeogenesis, glyoxylate cycle, glycogen metabolism), and amino acid/nitrogen metabolism (Fig. S2B; Table S1). Quantitative PCR (qPCR) analysis confirmed galactose - and Pbs2p-dependent induction of HOG targets, which included established targets of the HOG pathway (STL1, ENA1, GPD1, CTT1, and HSP12) to varying levels corresponding to the RNA seq analysis (Fig. 1D, [23], [24], [41], [72]). Likewise, the HOG pathway reporter p8XCRE-lacZ [73] was induced by galactose (by 2.3-fold) in a Pbs2p-dependent manner (Fig. 1E; salt induced the reporter by 3-fold). Therefore, the HOG pathway induces a transcriptional response during growth in galactose.
The filamentous growth pathway is induced during growth in galactose [16], [74], and cells undergo filamentous growth in this setting. The filamentous growth pathway shares components with the HOG pathway. Comparative RNA seq between wild-type cells and the ste12Δ mutant showed that the HOG and filamentous growth pathways induce different target genes. Known targets of the filamentous growth pathway were identified (FLO11, CLN1, PGU1, YLRO42C, BAR1, MSB2, Table S1, [11], [22], [27]) as well as new targets, including genes that regulate progression through the G2/M phase of the cell cycle (CLB1, CLB2 and SWE1, [75], [76]; Fig. 1F), bud-site-selection (Fig. 1F, BUD8, RAX2 and RSR1, [77]–[79]), a PM regulator of the PKC pathway (Fig. 1F, WSC2 [80]), and components of the filamentous growth pathway (Fig. 1F, SHO1 and TEC1 [12], [23], [81]), possibly leading to positive feedback [82]. These genes were not Pbs2p-dependent, and the filamentous growth pathway did not show induction of HOG pathway targets (Table S1). Therefore, the HOG and filamentous growth pathways mediate non-overlapping responses during growth on galactose.
Comparing HOG Pathway Activation by Galactose and Osmotic Stress
To further examine HOG pathway activation during growth in galactose, phosphorylation of the MAPK Hog1p (P∼Hog1p) was measured, which provides a readout of HOG pathway activity [73], [83]. This assay had the advantage of evaluating the kinetics and genetic pathways required for pathway activation. Consistent with the RNA seq analysis, growth of cells in galactose induced the activation of the HOG (Fig. 2A, P∼Hog1p) and filamentous growth pathways (Fig. 2A, P∼Kss1p). Depending on the condition, different levels of basal P∼Hog1p and P∼Kss1p were detected, which may represent differences in baseline activity between the pathways.
Fig. 2. Comparison of HOG pathway activation by galactose and osmotic stress. 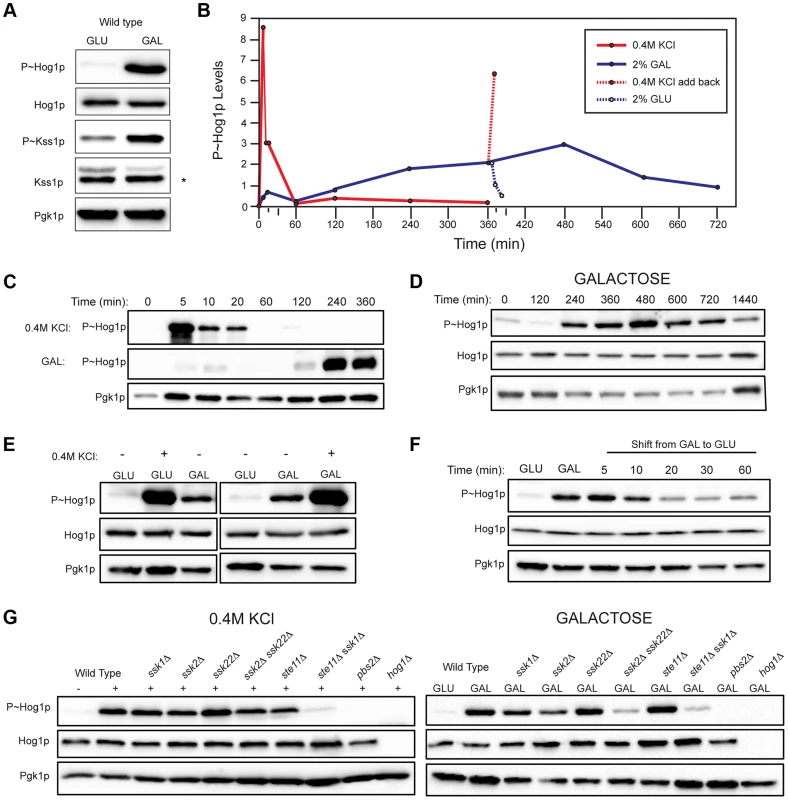
For all phosphoblots involving Hog1p and Kss1p, the sizes of proteins are P∼Hog1p (∼49 kDa), Hog1p (∼49 kDa), P∼Kss1p (∼43 kDa), Kss1p (∼43 kDa), and Pgk1p (∼45 kDa). Pgk1p was used as a loading control. Asterisk (*) refers to a background band detected by the Kss1p antibody. Basal P∼Hog1p and P∼Kss1p showed variable levels under un-inducing conditions. A) Wild type cells (PC538) cells were grown to mid-log phase (∼5.5 hrs) in YEPD (GLU) or YEP-GAL (GAL) media and evaluated by immunoblot analysis for phosphorylation of the MAPKs Hog1p and Kss1p. B) Graph of P∼Hog1p levels under the indicated conditions, as determined by ImageJ analysis. C) Time-course analysis. Wild-type cells (PC538) were grown to mid-log phase and transferred to media containing salt (YEPD+0.4 M KCl) or galactose (YEP-GAL) for the indicated times. D) Extended time course of Hog1∼P during growth in galactose. E) Combinatorial analysis of the response to osmotic stress and galactose. Cells were grown to mid-log phase in YEPD, YEP-GAL, or YEPD+0.4M KCl, which was added to the cells growing in YEPD for 5 min. F) P∼Hog1p levels in cells shifted from galactose (YEP-GAL) to glucose (YEPD) for the indicated time points. Cells in YEP-GAL media were harvested by centrifugation, washed twice in water, and resuspended in YEPD for the indicated time points. G) P∼Hog1p levels during growth in 0.4M KCl and galactose in mutants lacking Ssk1p or Ste11p branches of the HOG pathway. Wild type cells (PC538), and the ssk1Δ (PC1523), ssk2Δ (PC6086), ssk22Δ (PC6085), ssk2Δ ssk22Δ (PC6031), ste11Δ (PC3861), ste11Δ ssk1Δ (PC2061), pbs2Δ (PC2053) and hog1Δ (PC6047) mutants were grown in YEP-GAL medium or YEPD medium containing 0.4M KCl for 5 min. Other nutrient-limiting conditions were also tested. Non-fermentable carbon sources, acetate and ethanol-glycerol (Fig. S3A), and limitation of environmental nitrogen (Fig. S3B), also activated the HOG pathway. Other poor nutrients, like raffinose (Fig. S3C), limiting glucose (Fig. S3C), and phosphate limitation (Fig. S3D) did not induce the HOG pathway, which indicates that a particular metabolic context evokes the response (see below). Because galactose caused robust induction of the HOG and filamentous growth pathways, that inducer was used for subsequent experiments.
An established trigger of the HOG pathway is an increase in external osmolarity [84]. HOG pathway activation by high osmolarity (KCl, 0.4 M) and galactose (2%) was compared. In response to osmotic stress, P∼Hog1p was detected by 5 min. By 60 min, P∼Hog1p was reduced due to pathway attenuation (Fig. 2, B and C, upper panel [84], [85]). By comparison, P∼Hog1p was detected during growth in galactose at 240 min (Fig. 2, B and C, middle panel), and the signal persisted until 720 min (Fig. 2D). Salt (0.4 M KCl) induced higher levels of P∼Hog1p than growth in galactose (Fig. 2E), which is consistent with the RNA seq analysis (Table S1). Salt (0.4 M KCl) added to cells grown in galactose caused a rapid increase in P∼Hog1p (Fig. 2, B and E). Transfer of cells from galactose (YEP-GAL) to glucose (YEPD), which leads to glucose repression (see below), caused a reduction in P∼Hog1p levels by 20 min (Fig. 2, B and F), which was comparable to the reduction in P∼Hog1p levels in response to osmotic shock (Fig. 2, B and C). Therefore, the amplitude and duration of HOG pathway signaling differs depending on whether the inducer is osmotic stress or galactose.
Two branches of the HOG pathway regulate the response to osmotic stress, which converge on the MAPKK Pbs2p (Fig. S1, [13], [18], [19]). In response to salt, the branches are redundant, in that mutants lacking both branches (ssk1Δ ste11Δ) show the same defect as mutants lacking the MAPKK (pbs2Δ) or MAPK (hog1Δ, Fig. 2G). During growth in galactose, but not salt, the MAPKKKs Ssk2p and Ssk22p were required for HOG pathway activation (ssk2Δ ssk22Δ; Fig. 2G). Thus, Ssk2p and Ssk22p have a role in the response to galactose that differs from their role in response to osmotic stress. The different branches of the HOG pathway play different roles under different conditions, such as in the response to different concentrations of salt [23]. Nitrogen limitation showed a similar requirement for Ssk2p and Ssk22p (Fig. S3E). The Ste11p branch alone was not required (ste11Δ, Fig. 2G), but when the ste11Δ mutant was combined with the ssk1Δ mutant, a defect was observed (ssk1Δ ste11Δ, Fig. 2G). Thus, the Ste11p branch plays a minor role in the HOG response to galactose. One might expect that the Msb2p/Sho1p branch, which induces the filamentous growth pathway in response nutrient limitation, would transmit nutrient signals to Pbs2p/Hog1p (Fig. S1). In fact, the Sln1p branch played the major role in this nutritional response, whereas the Sho1p branch played a minor role.
The AMPK Snf1p and the TCA Cycle Are Required for HOG Pathway Activation by Galactose
Glucose is the preferred carbon source in yeast [86], [87]. When glucose is abundant, yeast cells exclusively utilize that nutrient over non-preferred carbon sources like galactose. Glucose repression prevents the transport and utilization of other carbon sources [86], [88]–[90]. As shown above, glucose added to cells grown on galactose resulted in attenuation of the HOG response (Fig. 2F). To further test whether glucose prevents the HOG response to galactose, cells were grown in media containing both glucose and galactose as a carbon source. Under this condition, HOG pathway signaling was also attenuated (Fig. 3A). These experiments indicate that galactose metabolism is required for HOG pathway activation. Consistent with this possibility, mutants defective for galactose transport and utilization (Fig. 3B; gal3Δ, gal4Δ, gal7Δ, and gal10Δ [88], [90]–[93]) were defective for HOG pathway activation.
Fig. 3. Role of increased metabolic respiration and Snf1p in activation of the HOG pathway. 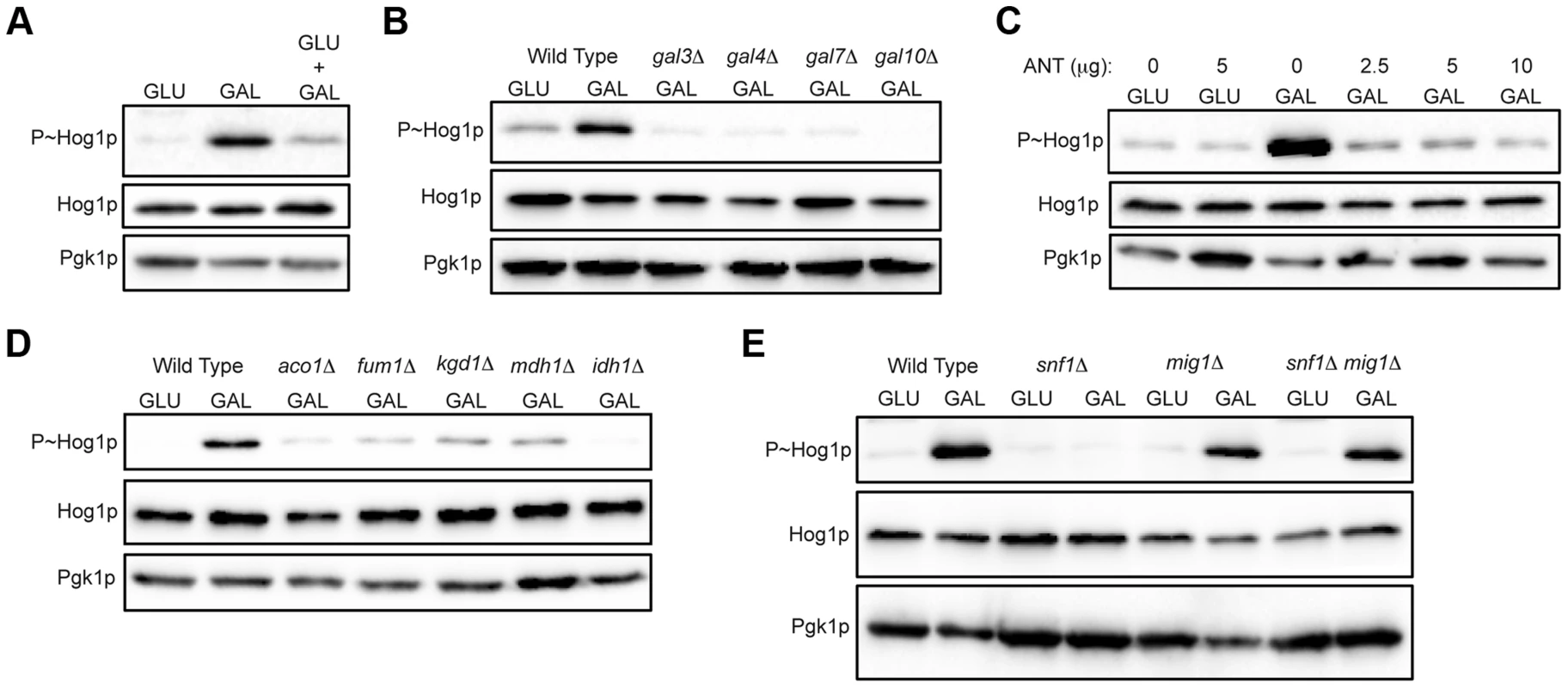
A) Immunoblot showing P∼Hog1p levels in cells grown in glucose (YEPD), galactose (YEP-GAL) or glucose and galactose (YEPD+2% GAL). B) Wild-type cells (PC6016) and the gal3Δ, gal4Δ, gal7Δ and gal10Δ mutants grown in YEP-GAL. C) P∼Hog1 levels in cells grown under the indicated conditions for 3 h with or without antimycin, ANT. D) Wild type (PC538) and the aco1Δ (PC3912), fum1Δ (PC6152), mdh1Δ (PC6153) and kgd1Δ (PC6155) and idh1Δ (PC6154) mutants were grown in galactose for 5.5 hrs. E) Wild-type cells (PC538), and the snf1Δ (PC560), mig1Δ (PC4843) and snf1Δ mig1Δ (PC6076) mutants were grown in YEP-GAL medium for 5.5 hrs. Galactose utilization increases the respiratory capacity by shunting ATP production through the electron transport chain [86], [94], [95]. An inhibitor of respiration, antimycin [96]–[98], prevented HOG pathway activation by galactose (ANT, Fig. 3C). Antimycin did not prevent HOG pathway activation in response to salt (Fig. S4A). Metabolic respiration produces intermediates that are utilized by the tricarboxylic acid (TCA, or citric acid) cycle to generate precursors for electron transport. Strains lacking TCA cycle enzymes aconitase (aco1Δ), fumarase (fum1Δ), malate dehydrogenase (mdh1Δ), alpha keto-glutarate (kgd1Δ) and iso-citrate dehydrogenase (idh1Δ) were defective for HOG pathway activation by galactose (Fig. 3D). These mutants were not required to mediate an osmotic response (Fig. S4B, shown for aco1Δ). Therefore, metabolic respiration of galactose underlies activation of the HOG pathway.
The AMP-dependent protein kinase (AMPK) Snf1p is a major regulator of the response to poor carbon source utilization [99], [100]. The main function of Snf1p is the de-repression of glucose-repressed genes [86], [89], [99]. Snf1p was required for HOG pathway activation by galactose (Fig. 3E). Snf1p functions with the regulatory subunit Snf4p [101], which was also required for HOG pathway activation by galactose (Fig. S4C). Snf1p and Snf4p were not required for HOG pathway activation in response to osmotic stress (Fig. S4D). Snf1p phosphorylates the transcriptional repressor Mig1p to relieve glucose repression [102]–[104], which leads to induction of the GAL genes and other genes [89], [102], [105], [106]. Loss of Mig1p restored HOG pathway activity in the snf1Δ mutant (Fig. 3E, mig1Δ snf1Δ). Cells lacking Mig1p alone did not influence HOG pathway activity (Fig. 3E, mig1Δ). Therefore, Snf1p regulates the HOG pathway through its major role in relieving de-repression of glucose-repressed genes. Snf1p also regulates nitrogen assimilation pathways [107] but was not required to activate the HOG pathway in response to nitrogen deficiency (Fig. S4E). In summary, metabolic respiration triggers AMPK-dependent activation of the HOG pathway.
UPR Kinase Ire1p Mediates HOG Pathway Activation by Galactose
Protein glycosylation is an oligosaccharide modification of proteins that occurs in the endoplasmic reticulum (ER) and Golgi apparatus [108]. Defects in protein glycosylation trigger a global response that involves the action of several MAPK pathways, including the filamentous growth [10], [60] and HOG pathways [63], [64]. Comparative RNA seq analysis identified HOG pathway targets induced by treatment with tunicamycin, an inhibitor of N-linked glycosylation (Fig. 1, B and C; Table S1). To further explore the HOG and filamentous growth pathway response to glycosylation deficiency, a conditional mutant, pmi40-101 [60], was used that is defective for an early step in N - and O-linked glycosylation [109], [110]. Growth of the pmi40-101 mutant in media lacking mannose induces its glycosylation defect and showed elevated HOG and filamentous growth pathway activity (Fig. 4A). Defects in O-linked glycosylation also modestly activated the HOG pathway (Fig. S5A). In response to glycosylation deficiency, HOG pathway activation did not require Snf1p (Fig. S5B), which is consistent with the idea that Snf1p regulates the HOG pathway by the de-repression of glucose-repressed genes.
Fig. 4. Role of the UPR in mediating HOG pathway activation during growth in galactose and in response to protein glycosylation deficiency. 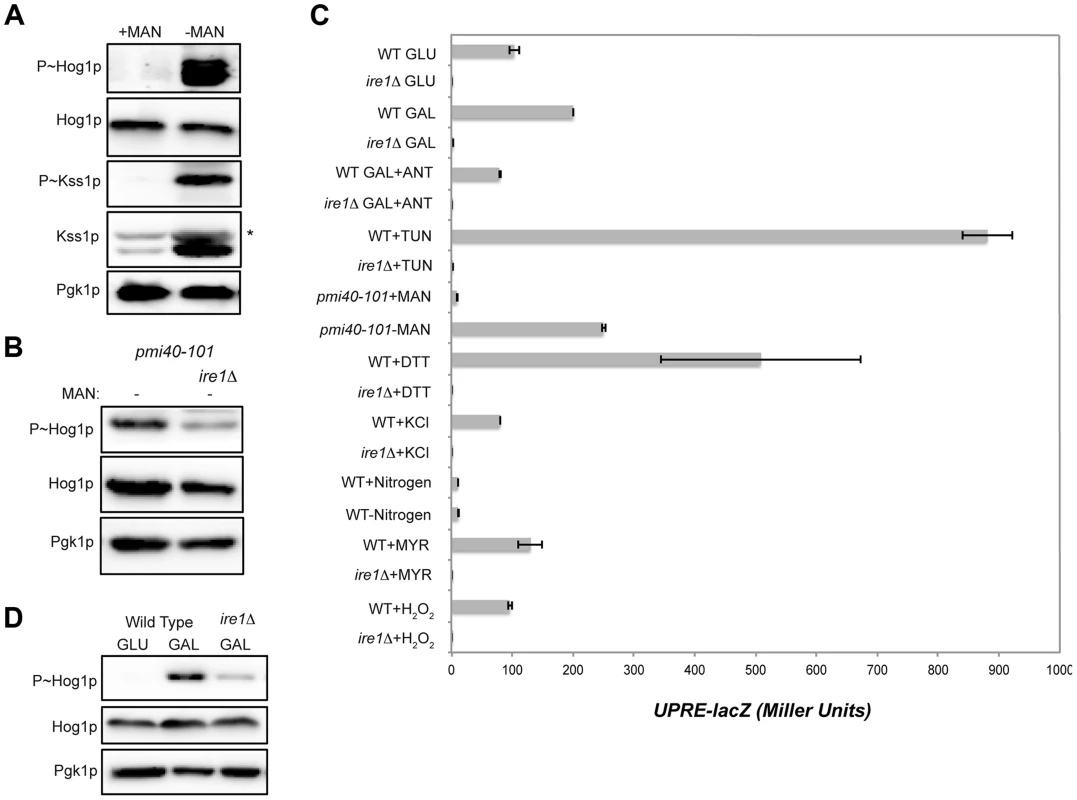
A) P∼Hog1p and P∼Kss1p levels in the pmi40-101 mutant (PC244) grown in YEPD+/−50 mM MAN (mannose) for 5.5 hrs. B) The pmi40-101 (PC244) and pmi40-101 ire1Δ (PC6044) mutants were grown in YEPD medium +/−50 mM MAN. C) Activity of the UPRE-lacZ reporter under the indicated conditions. H2O2 (5 mM; 3 hr), DTT (4 mM; 3 hr), KCl (1M; 30 min), TUN (2.5 µg; 3 hr), galactose (2%, 5.5 hr), MYR (myriocin) (5 µg; 3 hr), SD (+/−nitrogen; 5.5 hr), pmi40-101 (+/− MAN; 5.5 hr), ANT (2.5 µg; 3 hr). D) P∼Hog1p levels in the ire1Δ mutant grown in galactose. Wild-type cells (PC538) and the ire1Δ mutant (PC6048) were grown in YEP-GAL medium. Defects in protein glycosylation induce problems with protein folding in the ER, which activates the UPR [111], [112]. The UPR is regulated by the kinase Ire1p [113], which has recently been shown to mediate the HOG pathway response to glycosylation deficiency (Fig. 4B [63], [64]). As expected from these reports, Ire1p was required to mediate the HOG response to glycosylation deficiency (Fig. 4B). Protein glycosylation deficiency, as induced by tunicamycin or the pmi40-101 mutant, also induced a transcriptional reporter of the UPR (UPRE-lacZ, [113]) in an Ire1p-dependent manner (Fig. 4C).
An increase in metabolic respiration might also trigger ER stress that leads to Ire1p-dependent activation of the HOG pathway. HOG pathway activation in response to galactose was reduced in the ire1Δ mutant (Fig. 4D). Galactose also induced expression of the UPRE-lacZ reporter in an Ire1p-dependent manner (Fig. 4C). Induction of the UPRE-lacZ reporter by galactose was abolished by treatment with antimycin (Fig. 4C). Therefore, an increase in metabolic respiration stimulates the UPR, which leads to Ire1p-dependent activation of the HOG pathway.
The HOG pathway is activated by several stimuli, including salt, nitrogen (this study), myriocin [14], [114], and oxidative stress [115], [116]. The UPR was not induced by these stimuli (Fig. 4C). Therefore, two different types of inducers activate the HOG pathway. One type is Ire1p-dependent (induced by increased metabolic respiration and glycosylation deficiency), and another is Ire1p-independent (induced by salt and other stresses).
Filamentous Growth and HOG Pathways Contribute to Growth in Galactose and Modulate Each Other's Activities to Produce an Optimal Response
The filamentous growth and HOG pathways are activated during growth in galactose (Fig. 1A; Fig. 2A; Table S1). However, the HOG pathway inhibits the filamentous growth pathway in response to osmotic stress [12], [16], [47]–[49]. To determine whether the HOG pathway inhibits the filamentous growth pathway during growth in galactose, cells were examined by microscopy. Cells lacking an intact HOG pathway showed hyper-polarized growth (Fig. 5A, pbs2Δ), indicative of a hyperactive filamentous growth pathway. Comparative RNA seq showed that transcriptional targets of the filamentous growth pathway (PGU1, SVS1, MSB2, and KSS1; Table S1) were up-regulated in the pbs2Δ mutant in galactose. In line with the RNA seq data, the pbs2Δ mutant showed elevated activity of a filamentous growth pathway reporter (Fig. 5B, FRE-lacZ). Tyrosine phosphatases Ptp2p and Ptp3 negatively regulate the Hog1p pathway [85], [117]. The ptp2Δ ptp3Δ double mutant showed elevated P∼Hog1p levels and correspondingly lower levels of P∼Kss1p under pathway inducing conditions (galactose) (Fig. 5C). This was also observed under basal conditions (glucose) (Fig. 5C). Accordingly, the ptp2Δ ptp3Δ double mutant (and the ptp3Δ single mutant) showed reduced invasive growth and crosstalk reporter activity (Fig. S6). Therefore, the HOG pathway inhibits the filamentous growth pathway during growth in galactose.
Fig. 5. Cross-inhibition between the filamentous growth and HOG pathways during growth in galactose. 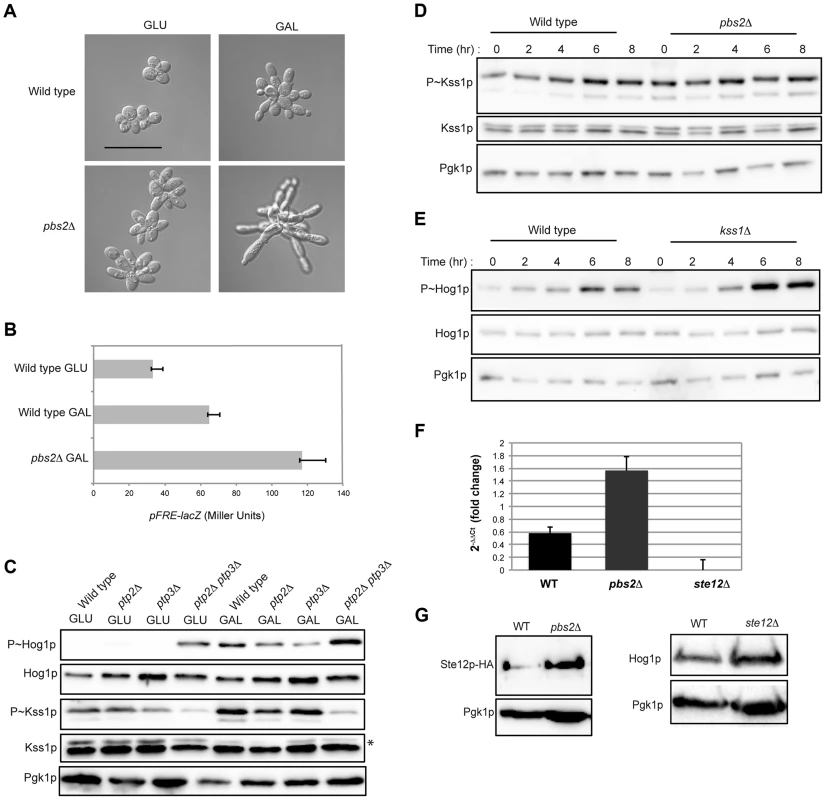
A) Morphology of wild-type cells (PC538) and the pbs2Δ mutant (PC2053), grown on YEPD and YEP-GAL for 24 hrs. Bar, 5 microns. B) pFRE-lacZ reporter activity in wild-type cells (PC313) and the pbs2Δ mutant (PC5035) in YEP-GAL medium. C) Role of protein tyrosine phosphatases in P∼Hog1p activity in galactose. Wild-type cells (PC538), and the ptp2Δ (PC6156), ptp3Δ (PC6157) and ptp2Δ ptp3Δ double mutant (PC6158) were grown in YEPD and YEP-GAL media for 5.5 hrs. D) P∼Kss1p activity in wild-type cells and the pbs2Δ mutant (PC2053) grown in YEP-GAL medium over a time course as indicated. E) P∼Hog1p activity in the kss1Δ mutant (PC620) grown in YEP-GAL medium for the times indicated. F) qPCR showing the relative expression of STE12 mRNA in the wild-type (PC538), pbs2Δ (PC2053) and ste12Δ (PC2382) mutant cells. Error bars indicate +/− standard error mean of three independent experiments. Actin (ACT1) mRNA was used as a control. G) Ste12p-HA protein levels in the wild-type and pbs2Δ strains. Hog1p levels by immunoblot analysis are also shown. The filamentous growth pathway also inhibits the HOG pathway [10]. Consistent with this finding, the level of P∼Kss1p was elevated in the pbs2Δ mutant at 0 h, 2 h, and 4 h (Fig. 5D) of growth in galactose. Similarly, the level of P∼Hog1p was elevated in the kss1Δ mutant at 6 h and 8 h (Fig. 5E). RNA seq analysis showed STE12 was up-regulated by galactose in the pbs2Δ mutant (Table S1). This was confirmed by qPCR (1.52 log2 fold) (Fig. 5F) and was reflected at the protein level (Fig. 5G). Likewise, Hog1p protein levels were modestly affected in the ste12Δ mutant (Fig. 5G). Thus, the HOG pathway inhibits the filamentous growth pathway during growth in galactose, which may occur at the STE12 level. These results show that the HOG and filamentous growth pathways modulate each other's activities in a complex pattern in galactose.To this point, our results suggest an apparent paradox. The HOG and filamentous growth pathways are both activated during growth on galactose, yet the pathways dampen each other's activities. To determine the roles of the pathways in this setting, mutants in the filamentous growth and HOG pathway pathways were examined for growth in galactose. Mutants lacking the filamentation MAPKK (ste7Δ) and HOG MAPKK (pbs2Δ) were not defective for growth on galactose (Fig. 6A). However, the ste7Δ pbs2Δ double mutant showed a growth defect (Fig. 6A). This defect was not specific for the MAPKKs, because another mutant that blocks the activity of both pathways showed an equivalent growth defect (ste11Δ ssk1Δ, Fig. 6A). The ste7Δ pbs2Δ double mutant also showed morphological defects during growth in galactose (Fig. 6B). Therefore, the HOG and filamentous growth pathways have a redundant function in promoting proper growth and morphogenesis during growth in galactose.
Fig. 6. Role of the HOG and filamentous growth pathways in growth in galactose and effect of the inhibitory role of the HOG pathway on filamentous growth pathway outputs. 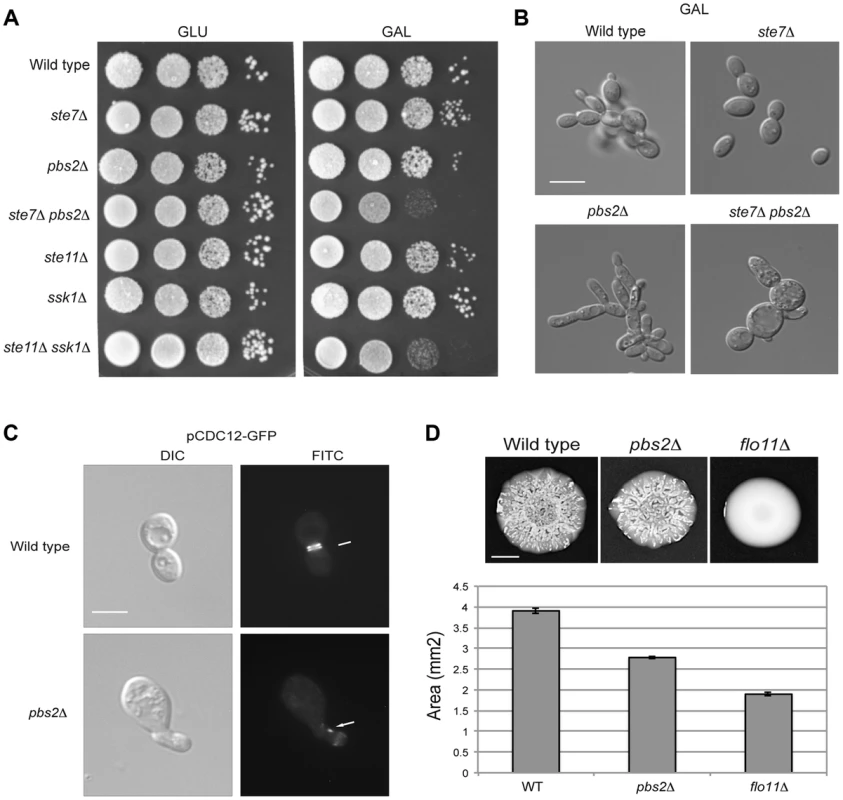
A) Serial dilutions of wild-type (PC313), ste7Δ (PC4928), pbs2Δ (PC5035) and ste7Δ pbs2Δ (PC6272) cells were spotted on YEPD and YEP-GAL media. B) Morphology of wild-type cells (PC538), the pbs2Δ mutant (PC2053), the ste7Δ mutant (PC4982), and the ste7Δ pbs2Δ double mutant (PC6272) grown YEP-GAL media for 24 hrs. Bar, 5 microns. C) Septin staining of wild-type and pbs2Δ cells harboring the pCdc12p-GFP plasmid. Cells were grown to mid-log phase in YEPD. D) Mat formation in cells lacking the filamentous growth or HOG pathways. Wild-type (PC538), flo11Δ (PC1029), and pbs2Δ (PC2053) strains were grown in YEPD medium for 16 hrs and then spotted onto low agar (0.3%) YEP-GAL medium for 3 d at 30°C. Bar, 1 cm. What is the benefit to the cross-modulation between the two pathways? One possibility is that modulation of the pathways' activities may be important to optimize the response. To test this possibility, the pbs2Δ mutant was examined in detail by microscopic examination. A subset of pbs2Δ cells showed morphological defects [∼5% compared to <0.5% of wild-type cells, >1000 cells counted (Fig. 6C)]. Thus, hyper-activation of the filamentous growth pathway can lead to morphogenetic defects. To further explore this possibility, the pattern of septins, which mark the mother-bud neck [118], [119], was also examined. Septin staining showed an irregular pattern pbs2Δ cells with morphological defects (Fig. 6C). This defect is indicative of problems with cell-cycle progression or proper growth.
To determine if the hyper-polarized growth of the pbs2Δ mutant comes from hyper-activation of the filamentous growth pathway, cell morphology was quantitated by microscopy. In rich media (YEPD), wild-type cells grow predominately in the vegetative (round) form (8+/−2% elongated cells; 200 cells counted for all trials). By comparison, the pbs2Δ mutant shows a cell-elongation morphology (>83.5+/−5% elongated cells) that was abolished in the pbs2Δ ste7Δ double mutant (7+/−3% elongated cells). Therefore, the enhanced polarized morphology seen in pbs2Δ cells, and concomitant morphological abnormalities, is due to Ste7p. This result is complicated because in YEP-GAL, the ste7Δ pbs2Δ double mutant shows morphological defects not seen in either single mutant (Fig. 6B). Thus, Pbs2p may attenuate morphogenesis in multiple ways.
This finding extends to other negative-regulatory inputs to the filamentous growth pathway as well (Chavel et al. IN PRESS). Although only a low percentage of cells exhibit morphological defects, it is likely that even minor mis-coordination of basic cellular processes would be detrimental to cell health. Thus, modulation of the filamentous growth pathway by the HOG pathway is necessary for proper cell growth and morphogenesis.
As a second test, the response of a population of cells was examined. Yeast cells expand in biofilms/mats through the action of the filamentous growth pathway, which regulates expression of the cell-adhesion molecule Flo11p [120]. When hyper-activated, the filamentous growth pathway causes an increase in FLO11 expression that prevents biofilm/mat expansion [121], [122]. We found that the pbs2Δ mutant formed smaller biofilms/mats than wild-type cells during expansion on galactose media (Fig. 6D, see quantitation in graph). Thus, modulation of the filamentous growth pathway by the HOG pathway is required to coordinate cell growth and optimize colonial behavioral responses.
The Ire1p-HOG1p-ERK-p38 Axis Is an Evolutionarily Conserved Response among Fungal Species
The signaling circuit characterized here might be specific to S. cerevisiae or extend to other species. To address this question, pathways of the fungal pathogen Candida albicans were examined. Like budding yeast, C. albicans has a Kss1p-type pathway (Cek1p pathway [123]–[125]), and a p38-type pathway (CaHOG pathway [126]). The CaHOG MAPK CaHog1p is activated by osmotic stress (Fig. 7A [127]) and was also induced by tunicamycin, myriocin, and growth in galactose (Fig. 7A, 5 h at 30°C). Thus, the versatility of HOG pathway in sensing diverse stresses is conserved among several fungal species.
Fig. 7. MAPK responses in C. albicans during growth in galactose. 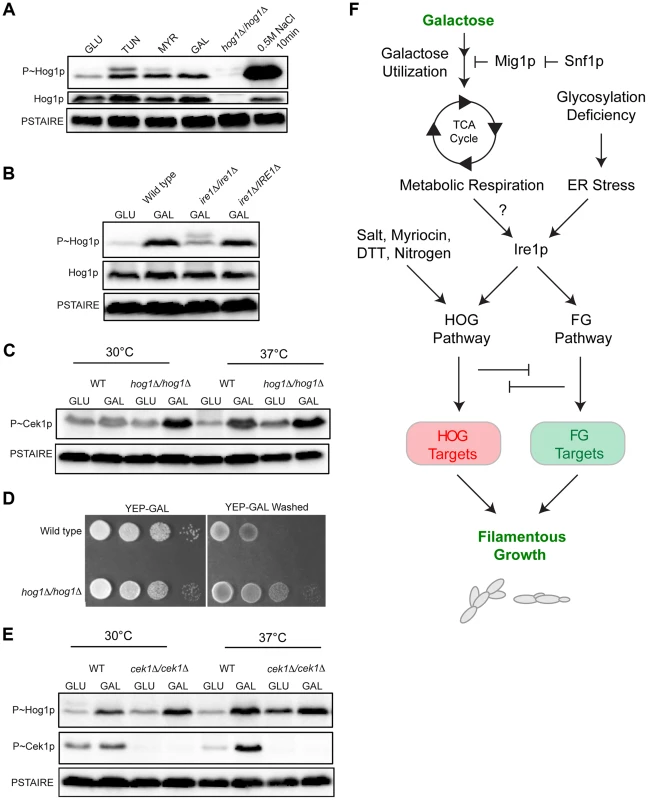
A) Immunoblot analysis of P∼CaHog1p. Wild-type (PC6111) cells were grown in YEPD and YEP-GAL medium (5.5 hrs) and treated with TUN (tunicamycin) (2.5 µg for 3 hrs), MYR (myriocin) (2.5 µg for 3 hrs) and 0.5M NaCl (10 min). B) Phosphorylation of CaHog1p requires the CaIre1p. Wild-type (PC6116), ire1Δ/ire1Δ (PC6144), and ire1Δ/pIRE1 (PC6145) cells were grown in YEPD and YEP-GAL medium (5.5 hrs). C) P∼Cek1p levels in the hog1Δ/hog1Δ mutant at 30°C and 37°C. Wild-type (PC6111) and hog1Δ/hog1Δ (PC5008) cells were grown in YEPD and YEP-GAL medium (5.5 hrs). D) Plate-washing assay of wild-type cells (PC6111) and the hog1Δ/hog1Δ mutant (PC5008) on YEP-GAL medium at 37°C for 48 hrs. The plate was photographed, washed, and photographed again to reveal invaded cells. E) P∼CaHog1p and P∼Cek1p levels in the cek1Δ/cek1Δ mutant at 30°C and 37°C. Wild-type (PC6111) and cek1Δ/cek1Δ (PC6114) cells were grown in YEPD and YEP-GAL medium (5.5 hrs). F) Model showing the roles of the HOG and filamentous growth pathways in the response to growth in galactose. Galactose is transported into cells and metabolized by genes under the control of Snf1p. As a result, metabolic respiration is increased, which by some mechanism (?) induces the UPR. Ire1p mediates activation of the HOG and filamentous growth pathways (Adhikari et al. SUBMITTED). The HOG and filamentous growth pathways induce different target genes to redundantly promote growth under this condition. The antagonistic roles of these pathways on each other's activities optimize the response. We also tested whether CaIre1p is required to mediate the HOG pathway response to galactose. The ire1Δ/ire1Δ double mutant was defective for producing the elevated levels of P∼CaHog1p seen during galactose treatment in wild-type cells (Fig. 7B). A strain lacking CaIre1p but containing a complemented version ire1Δ/IRE1, restored P∼CaHog1p activity (Fig. 7B). Thus, CaIre1p mediates the HOG pathway response to galactose.
Previous reports have shown that the C. albicans HOG pathway negatively regulates the Cek1p pathway [128]. In C. albicans, growth at high temperatures (37°C) is a potent inducer of dimorphism [129], [130]. Interestingly, growth of C. albicans cells in galactose induced P∼Cek1p levels only at 37°C in wild-type cells (Fig. 7C). The C. albicans hog1Δ/hog1Δ mutant grown in galactose showed elevated P∼Cek1p levels at 30°C and 37°C (Fig. 7C). Similarly, the hog1Δ/hog1Δ mutant showed hyper-invasive growth compared to wild-type cells (Fig. 7D). Thus, CaHog1p inhibits Cek1p pathway activity during growth in galactose.
The Cek1p pathway might also inhibit the CaHOG pathway. In the cek1Δ/cek1Δ mutant, elevated P∼CaHog1p levels were observed during growth in galactose at 30°C and 37°C (Fig. 7E). Therefore, the CaHog1p pathway is activated by galactose in a Ire1p-dependent manner. Under this condition, the CaHog1p pathway and Cek1p pathways are both activated and both modulate each other's activities. These results indicate that the signaling axis described in S.cerevisiae extends to the opportunistic pathogen C. albicans.
Discussion
Differentiation into specialized cell types occurs during development and in response to extrinsic cues. Fungal species differentiate into the filamentous/hyphal cell type, which in pathogens occurs during colonization of the host. Using a genomic survey, RNA seq analysis, we identify a new role for a p38 MAPK pathway (HOG) in differentiation to the filamentous cell type in yeast. The HOG pathway is activated during growth in poor carbon sources through a regulatory circuit involving the AMPK Snf1p. Since deletion of MIG1 alleviates the need for Snf1p, the regulation of Hog1p is likely to be downstream of Mig1p repressed genes. HOG pathway activation in this context also required the ER stress kinase Ire1p. The connection between Ire1p and the HOG pathway has been reported [63], [64]. Our study therefore connects AMPK and Ire1p in a regulatory circuit that governs p38 (Fig. 7F). The HOG pathway and the ERK-type filamentous growth pathway induced target genes to promote growth in galactose, but the pathways also modulated each other's activities. Such modulation optimized cell growth and morphogenesis to facilitate production of the filamentous cell type (Fig. 7F). Our findings therefore elucidate a signaling network that occurs during differentiation and highlights the critical role for pathway modulation in proper cell-type specification.
Nutrient sensing in yeast has been extensively studied. Well-established pathways mediate the response to carbon and nitrogen limitation. We show that limiting nutrients, like non-preferred carbon and nitrogen sources, activate the p38-type HOG pathway in yeast. As a result, glucose repression, the AMPK Snf1p, metabolic respiration, and the TCA cycle feed into HOG pathway signaling (Fig. 7F). The HOG pathway is well known for its ability to sense and respond to changes in external osmolarity. Multiple inducers activate the HOG pathway, including citric acid [131], hypoxia [132], cold stress [133] and defective sphingolipid biosynthesis [114]. Here we demonstrate a connection between nutrients and activation of the HOG pathway (Fig. 7F). In mammals, p38-type stress activated protein kinase (SAPK) pathways mediate AMPK-dependent metabolic reprogramming. The p38 pathway can alter the balance between survival and apoptosis [134]. p38 also regulates respiration in muscles, and gluconeogenesis in liver, and when mis-regulated can lead to problems that range from diabetes [135]–[137] to tumor malignancy [138].
We show that in yeast, metabolic respiration feeds into the HOG response and requires the UPR regulator Ire1p. Several inducers of the Ire1p have been identified [139] but to our knowledge, this is the first example of a connection between defects in metabolic respiration and Ire1p (Fig. 7F). Hints at this connection come from studies in mammalian cells. Ire1p can regulate AMPK function [140], and glucose levels are connected in some manner to Ire1p activity [141]. Elements of the ER stress pathway drive metabolic reprogramming in triple-negative breast cancer cells [142]. In tumor microenvironments, Ire1p is required to promote the balance between lipid and protein biosynthesis potentially at the level of ER production [143]. It is plausible that Ire1p regulates p38 activity in these settings as well. It is unlikely that the AMPK-Ire1p-p38 circuit governs all p38-type responses; however, the connections reported here may underlie nutrient-dependent p38-type responses in many settings.
Metabolic respiration (and defects in protein glycosylation) induce the HOG (p38) and filamentous growth (ERK) pathways. The two pathways do not depend on each other for activation, and they induce non-overlapping targets (herein and [23], [82]). Both pathways are redundant for full growth on galactose, and the two pathways modulate each other's activities. This complicated functional interplay between the pathways is critical for proper cell growth and optimal differentiation to the filamentous cell type. Generally speaking, p38 and ERK pathways can be activated in the same cells to together orchestrate complex responses that include cell differentiation [144], [145]. An important insight from our study is that p38 and ERK pathways modulate each other's activities to produce an optimal response. Such cross-wiring to fine tune responses underscores the importance of precision in cell differentiation.
Materials and Methods
Strains and Plasmids and Growth Conditions
Yeast and bacterial strains were grown by standard methods [146], [147]. Yeast strains were grown on YEP (yeast extract and peptone) medium containing 2% glucose (D) or 2% galactose (GAL) unless otherwise indicated. Cells were grown at 30°C. Strains are listed in Table 1. Primers used in the study are listed in Table 2. The plate-washing assay [148] and the single cell invasive growth assay [60] were performed as described. Biofilm assays were performed as described [120], except that galactose was used as a carbon source.
Tab. 1. Strains used in this study. 
All strains are in the Σ1278b background unless otherwise indicated. Tab. 2. Primers used for qPCR in the study. 
Plasmid YCp-Cdc12p-GFP was provided by J. Pringle [149], pFRE-lacZ by H. Madhani [150], pUPRE-lacZ by David Eide [151], and p8X-CRE-lacZ by H. Saito [73]. Plasmid selection was maintained in synthetic complete medium containing 2% glucose (SD) or 2% galactose (S-GAL) that lacked uracil (-URA) or leucine (-LEU). Gene disruptions were performed according to standard genetic techniques [152], [153].
RNA Preparation
Total RNA was isolated by acid phenol method from 10 ml cultures of WT and pbs2Δ mutant grown in YEPD (5.5 hrs), YEP-GAL (5.5 hrs), YEPD+Tunicamycin (3 hrs) and YEPD+salt (10 min). Isolated RNA was purified over a RNAeasy column (Qiagen). RNA concentration was measured using the NanoDrop2000 spectrophotometer (Thermo Scientific, Wilmington, DE) and purity was established by the A260/A280 ratio. RNA was detected by running samples on an 8M Urea 6%polyacylamide gel, stained by ethidium bromide. Three independent inductions were evaluated for RNA-seq analysis. Total RNA integrity was checked using an Agilent 2200 TapeStation (Agilent Technologies, Inc., Santa Clara, CA) and quantified using a Trinean DropSense96 spectrophotometer (Caliper Life Sciences, Hopkinton, MA).
RNA-seq Expression Analysis
RNA sequencing was performed at the Fred Hutchinson Cancer Research Center (Seattle, WA). RNA seq was performed in triplicate by sequencing RNA prepared from 3 different (independent) cultures. RNA-seq libraries were prepared from total RNA using the TruSeq RNA Sample Prep Kit (Illumina, Inc., San Diego, CA, USA). Library size distributions were validated using an Agilent 2200 TapeStation (Agilent Technologies, Santa Clara, CA, USA). Additional library QC, blending of pooled indexed libraries, and cluster optimization was performed using Life Technologies' Invitrogen Qubit 2.0 Fluorometer (Life Technologies-Invitrogen, Carlsbad, CA, USA). RNA-seq libraries were pooled (24-plex) and clustered onto a flow cell lane using an Illumina cBot. Sequencing was performed using an Illumina HiSeq 2500 in Rapid Mode employing a paired-end, 50 base read length (PE50) sequencing strategy.
Image analysis and base calling were performed using Illumina's Real Time Analysis v1.17 software, followed by ‘demultiplexing’ of indexed reads and generation of FASTQ files, using Illumina's CASAVA v1.8.2 software (http://www.illumina.com/software.ilmn). For analysis of the RNA seq data, reads of low quality were filtered out prior to alignment to the reference genome (S. cerevisiae assembly R64-1-1, Ensembl release 75) using TopHat v2.0.9 [154]. Counts were generated from TopHat alignments for each gene using the Python package HTSeq v0.5.4 (http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html). Genes with low counts across all samples were removed, prior to identification of differentially expressed genes using the Bioconductor package edgeR v3.4.2 [155]. A false discovery rate (FDR) method was employed to correct for multiple testing [156]. Differential expression was defined as |log2 (ratio) |≥0.585 (±1.5-fold) with the FDR set to 5%.
Quantitative RT-PCR Analysis
cDNA was synthesized using iScript cDNA synthesis kit (BioRAD, Carlsbad CA) according to manufacturer's protocol. PCR reactions were set-up using iQ SYBR Green Supermix (BioRAD, Carlsbad, CA). qPCR was performed using the following amplification cycles: initial denaturation for 8 min at 95°C, followed by 35 cycles (denaturation for 15 sec at 95°C and annealing for 1 min at 60°C). Expression of genes was quantified using the 2−ΔΔCt method [157] where ACT1 (actin) was used for normalization of expression values.
Cell Inductions and Protein Immunoblot Analysis
To analyze HOG and filamentous growth pathway activity by phosphoblot analysis, cells were induced under the following conditions. For S. cerevisiae, cells were grown in YEPD to mid-log phase, and 0.4 M KCl was added for 5 min. For C. albicans, 0.4 M NaCl was used. For galactose, cells were grown in YEP-GAL medium to mid-log phase (5.5 hrs). For tunicamycin, cells were grown to mid-log phase in YEPD, and tunicamycin was added to cells for 3 hrs. [2.5 µg Sigma CAT. # T7765]. Antimycin [Sigma CAT # A8674] was added to mid-log phase cells for 3 hrs. (2.5 µg, 3 hrs), H2O2 [Sigma CAT #216763] was added to cells at a concentration of 5 mM for 20 min. Myriocin [Sigma CAT #M1177]. Yeast strains were grown in nitrogen free media (yeast nitrogen base without amino acids and without ammonium sulfate [1.7 g/L] BD Franklin Lakes, NJ; #233520) [31] supplemented with glucose as a carbon source. For phosphate free medium (yeast nitrogen base without amino acids and without phosphate [5.6 g/L], MP Biomedicals LLC, Solon, OH; #114027812) was supplemented with amino acids as a nitrogen source and glucose as a carbon source. For induction of Candida albicans pathways, strains were maintained at 30°C. Cells were grown to mid-log phase (∼5 hrs) in YEPD or YEP-GAL and treated with 0.5 M NaCl for 10 min, myriocin (5 mM for 10 min), and tunicamycin (2.5 µg for 3 hrs). The pmi40-101 mutant was grown in YEPD medium supplemented with or without 50 mM mannose for 5 hrs. For strains that exhibited growth defects in galactose, input cell number (OD600) was increased to be equivalent to wild-type cells at mid log phase.
Cell extracts were prepared for immunoblot analysis according to established procedures [158]. Mid-log phase cells were harvested by centrifugation, and proteins were precipitated by trichloroacetic acid [TCA]. Cells were lysed in the TCA buffer (10 mM Tris HCl pH 8.0; 10%TCA; 25 mM ammonium acetate; 1 mM EDTA) containing glass beads using FastPrep-24 Instrument (MP Biomedicals LLC, Solon, OH). After high-speed centrifugation the pellet was thoroughly mixed in the resuspension buffer (0.1M Tris HCl pH 11.0; 3%SDS) and boiled for 5 min and centrifuged for 30 sec at 16000 g. To the supernatant, equal volume of 2× SDS loading dye (100 mM Tris HCl pH 6.8; 4%SDS; 0.2% Bromophenol Blue; 20% glycerol; 200 mM ß-mercaptoethanol) was added.
Protein samples were separated on 10% SDS polyacrylamide gels (SDS-PAGE) and transferred to nitrocellulose membranes (Protran BA85, VWR International Inc., Bridgeport NJ). The membrane was blocked in immunoblot buffer (5% nonfat dry milk, 10 mM Tris-HCl [pH 8], 150 mM NaCl and 0.05% Tween 20) for 16 h at 4°C. WesternBright MCF fluorescent Western blotting kit from Advansta Inc. (Menlo Park, CA; LPS #K-12045-D20) was used for detection. Pgk1p antibodies (Life Technologies, Camarillo, CA; #459250) were used as a loading control. P∼Hog1p was detected using phospho-p38 antibodies (Cell Signaling Technology, Danvers, MA; #9211). S. cerevisiae Hog1p was detected by Hog1p antibodies (Santa Cruz Biotechnology, Santa Cruz, CA; #yC-20). C. albicans Hog1p was detected by (Santa Cruz Biotechnology, Santa Cruz, CA; #y-215). Cdc2 p34 antibody that recognizes PSTAIRE motifs in cyclin dependent kinases was used as a loading control for Candida protein extracts (Santa Cruz Biotechnology, Santa Cruz, CA; #sc-53). Phosphorylated Kss1p was detected by p42/p44 antibodies (Cell Signaling Technology, Danvers, MA; #4370) and total Kss1p was detected by (Santa Cruz Biotechnology, Santa Cruz, CA; #6775). Secondary antibodies, goat α-mouse IgG–HRP (Bio-Rad Laboratories, Hercules, CA; #170-6516), goat α-rabbit IgG-HRP (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA; #111-035-144), donkey α-goat IgG-HRP (Santa Cruz Biotechnology, Santa Cruz, CA; #sc-2020) were used and incubated for 1 hr at 20°C. Ponceau S (Sigma, St. Louis, MO; #P7170) was used to confirm equal loading among samples.
ß-Galactosidase Assays
ß-galactosidase assays were performed as described [11]. Cells were grown in selective media (SD-URA) for 16 hrs and sub-cultured in YEPD or YEP-GAL media for 5.5 hrs. Three independent experiments were performed and the average values are represented. Error bars indicate the standard deviation between trials.
Microscopy
Differential-interference-contrast (DIC) and fluorescence microscopy was performed with an Axioplan 2 fluorescent microscope (Zeiss) with a PLAN-APOCHROMAT 100×/1.4 (oil) objective (N.A. 0.17). Digital images were obtained with the Axiocam MRm camera (Zeiss). Image Acquisition and analysis was carried out using Axiovision 4.4 software (Zeiss).
Bioinformatics Analysis
Heat maps were generated using MeV (MultiExperiment Viewer) (http://www.tm4.org/mev.html). ImageJ analysis was used to quantitate band intensity for protein gels and immunoblots (http://imagej.nih.gov [159]) using the invert function and by subtraction of background signals. SGD was used for yeast gene annotation and analysis (http://www.yeastgenome.org). RNA seq data was evaluated and represented by Microsoft EXCEL software.
Supporting Information
Zdroje
1. NobileCJ, MitchellAP (2006) Genetics and genomics of Candida albicans biofilm formation. Cell Microbiol 8 : 1382–1391.
2. LengelerKB, DavidsonRC, D'SouzaC, HarashimaT, ShenWC, et al. (2000) Signal transduction cascades regulating fungal development and virulence. Microbiol Mol Biol Rev 64 : 746–785.
3. CullenPJ, SpragueGFJr (2012) The regulation of filamentous growth in yeast. Genetics 190 : 23–49.
4. BrucknerS, MoschHU (2012) Choosing the right lifestyle: adhesion and development in Saccharomyces cerevisiae. FEMS Microbiol Rev 36 : 25–58.
5. SaitoH, PosasF (2012) Response to hyperosmotic stress. Genetics 192 : 289–318.
6. HohmannS (2002) Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev 66 : 300–372.
7. SchwartzMA, MadhaniHD (2004) Principles of map kinase signaling specificity in Saccharomyces cerevisiae. Annu Rev Genet 38 : 725–748.
8. QiM, ElionEA (2005) MAP kinase pathways. J Cell Sci 118 : 3569–3572.
9. MurphyLO, BlenisJ (2006) MAPK signal specificity: the right place at the right time. Trends Biochem Sci 31 : 268–275.
10. YangHY, TatebayashiK, YamamotoK, SaitoH (2009) Glycosylation defects activate filamentous growth Kss1 MAPK and inhibit osmoregulatory Hog1 MAPK. Embo J 28 : 1380–1391.
11. CullenPJ, SabbaghWJr, GrahamE, IrickMM, van OldenEK, et al. (2004) A signaling mucin at the head of the Cdc42 - and MAPK-dependent filamentous growth pathway in yeast. Genes Dev 18 : 1695–1708.
12. O'RourkeSM, HerskowitzI (1998) The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev 12 : 2874–2886.
13. PosasF, Wurgler-MurphySM, MaedaT, WittenEA, ThaiTC, et al. (1996) Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell 86 : 865–875.
14. PosasF, SaitoH (1997) Osmotic activation of the HOG MAPK pathway via Ste11p MAPKKK: scaffold role of Pbs2p MAPKK. Science 276 : 1702–1705.
15. WuC, JansenG, ZhangJ, ThomasDY, WhitewayM (2006) Adaptor protein Ste50p links the Ste11p MEKK to the HOG pathway through plasma membrane association. Genes Dev 20 : 734–746.
16. PitoniakA, BirkayaB, DionneHM, VadaieN, CullenPJ (2009) The Signaling Mucins Msb2 and Hkr1 Differentially Regulate the Filamentation Mitogen-activated Protein Kinase Pathway and Contribute to a Multimodal Response. Molecular Biology of the Cell 20 : 3101–3114.
17. TatebayashiK, TanakaK, YangHY, YamamotoK, MatsushitaY, et al. (2007) Transmembrane mucins Hkr1 and Msb2 are putative osmosensors in the SHO1 branch of yeast HOG pathway. Embo J 26 : 3521–3533.
18. MaedaT, TakekawaM, SaitoH (1995) Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3-containing osmosensor. Science 269 : 554–558.
19. PosasF, SaitoH (1998) Activation of the yeast SSK2 MAP kinase kinase kinase by the SSK1 two-component response regulator. Embo J 17 : 1385–1394.
20. MaedaT, Wurgler-MurphySM, SaitoH (1994) A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369 : 242–245.
21. OtaIM, VarshavskyA (1993) A yeast protein similar to bacterial two-component regulators. Science 262 : 566–569.
22. RobertsCJ, NelsonB, MartonMJ, StoughtonR, MeyerMR, et al. (2000) Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science 287 : 873–880.
23. O'RourkeSM, HerskowitzI (2004) Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol Biol Cell 15 : 532–542.
24. PosasF, ChambersJR, HeymanJA, HoefflerJP, de NadalE, et al. (2000) The transcriptional response of yeast to saline stress. J Biol Chem 275 : 17249–17255.
25. McCleanMN, ModyA, BroachJR, RamanathanS (2007) Cross-talk and decision making in MAP kinase pathways. Nat Genet 39 : 409–414.
26. RuaD, TobeBT, KronSJ (2001) Cell cycle control of yeast filamentous growth. Curr Opin Microbiol 4 : 720–727.
27. MadhaniHD, GalitskiT, LanderES, FinkGR (1999) Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc Natl Acad Sci U S A 96 : 12530–12535.
28. KronSJ, StylesCA, FinkGR (1994) Symmetric cell division in pseudohyphae of the yeast Saccharomyces cerevisiae. Mol Biol Cell 5 : 1003–1022.
29. RuppS, SummersE, LoHJ, MadhaniH, FinkG (1999) MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. Embo J 18 : 1257–1269.
30. GuoB, StylesCA, FengQ, FinkGR (2000) A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc Natl Acad Sci U S A 97 : 12158–12163.
31. GimenoCJ, LjungdahlPO, StylesCA, FinkGR (1992) Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68 : 1077–1090.
32. CullenPJ, SpragueGFJr (2002) The roles of bud-site-selection proteins during haploid invasive growth in yeast. Mol Biol Cell 13 : 2990–3004.
33. TaheriN, KohlerT, BrausGH, MoschHU (2000) Asymmetrically localized Bud8p and Bud9p proteins control yeast cell polarity and development. Embo J 19 : 6686–6696.
34. BrewsterJL, GustinMC (1994) Positioning of cell growth and division after osmotic stress requires a MAP kinase pathway. Yeast 10 : 425–439.
35. BaltanasR, BushA, CoutoA, DurrieuL, HohmannS, et al. (2013) Pheromone-induced morphogenesis improves osmoadaptation capacity by activating the HOG MAPK pathway. Sci Signal 6: ra26.
36. WarringerJ, HultM, RegotS, PosasF, SunnerhagenP (2010) The HOG pathway dictates the short-term translational response after hyperosmotic shock. Mol Biol Cell 21 : 3080–3092.
37. TeigeM, ScheiklE, ReiserV, RuisH, AmmererG (2001) Rck2, a member of the calmodulin-protein kinase family, links protein synthesis to high osmolarity MAP kinase signaling in budding yeast. Proc Natl Acad Sci U S A 98 : 5625–5630.
38. Bilsland-MarchesanE, ArinoJ, SaitoH, SunnerhagenP, PosasF (2000) Rck2 kinase is a substrate for the osmotic stress-activated mitogen-activated protein kinase Hog1. Mol Cell Biol 20 : 3887–3895.
39. LeeJ, ReiterW, DohnalI, GregoriC, Beese-SimsS, et al. (2013) MAPK Hog1 closes the S. cerevisiae glycerol channel Fps1 by phosphorylating and displacing its positive regulators. Genes Dev 27 : 2590–2601.
40. Nadal-RibellesM, CondeN, FloresO, Gonzalez-VallinasJ, EyrasE, et al. (2012) Hog1 bypasses stress-mediated down-regulation of transcription by RNA polymerase II redistribution and chromatin remodeling. Genome Biol 13: R106.
41. De NadalE, ZapaterM, AlepuzPM, SumoyL, MasG, et al. (2004) The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature 427 : 370–374.
42. MasG, de NadalE, DechantR, Rodriguez de la ConcepcionML, LogieC, et al. (2009) Recruitment of a chromatin remodelling complex by the Hog1 MAP kinase to stress genes. EMBO J 28 : 326–336.
43. ZapaterM, SohrmannM, PeterM, PosasF, de NadalE (2007) Selective requirement for SAGA in Hog1-mediated gene expression depending on the severity of the external osmostress conditions. Mol Cell Biol 27 : 3900–3910.
44. ChowdhuryS, SmithKW, GustinMC (1992) Osmotic stress and the yeast cytoskeleton: phenotype-specific suppression of an actin mutation. J Cell Biol 118 : 561–571.
45. YuzyukT, AmbergDC (2003) Actin recovery and bud emergence in osmotically stressed cells requires the conserved actin interacting mitogen-activated protein kinase kinase kinase Ssk2p/MTK1 and the scaffold protein Spa2p. Mol Biol Cell 14 : 3013–3026.
46. YuzyukT, FoehrM, AmbergDC (2002) The MEK kinase Ssk2p promotes actin cytoskeleton recovery after osmotic stress. Mol Biol Cell 13 : 2869–2880.
47. ShockTR, ThompsonJ, YatesJR3rd, MadhaniHD (2009) Hog1 mitogen-activated protein kinase (MAPK) interrupts signal transduction between the Kss1 MAPK and the Tec1 transcription factor to maintain pathway specificity. Eukaryot Cell 8 : 606–616.
48. DavenportKD, WilliamsKE, UllmannBD, GustinMC (1999) Activation of the Saccharomyces cerevisiae filamentation/invasion pathway by osmotic stress in high-osmolarity glycogen pathway mutants. Genetics 153 : 1091–1103.
49. WestfallPJ, ThornerJ (2006) Analysis of mitogen-activated protein kinase signaling specificity in response to hyperosmotic stress: use of an analog-sensitive HOG1 allele. Eukaryot Cell 5 : 1215–1228.
50. LevinDE (2011) Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics 189 : 1145–1175.
51. Rodriguez-PenaJM, GarciaR, NombelaC, ArroyoJ (2010) The high-osmolarity glycerol (HOG) and cell wall integrity (CWI) signalling pathways interplay: a yeast dialogue between MAPK routes. Yeast 27 : 495–502.
52. GarciaR, Rodriguez-PenaJM, BermejoC, NombelaC, ArroyoJ (2009) The High Osmotic Response and Cell Wall Integrity Pathways Cooperate to Regulate Transcriptional Responses to Zymolyase-induced Cell Wall Stress in Saccharomyces cerevisiae. J Biol Chem 284 : 10901–10911.
53. BermejoC, RodriguezE, GarciaR, Rodriguez-PenaJM, Rodriguez de la ConcepcionML, et al. (2008) The sequential activation of the yeast HOG and SLT2 pathways is required for cell survival to cell wall stress. Mol Biol Cell 19 : 1113–1124.
54. DohlmanHG, ThornerJW (2001) Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu Rev Biochem 70 : 703–754.
55. ElionEA (2000) Pheromone response, mating and cell biology. Curr Opin Microbiol 3 : 573–581.
56. RagniE, PibergerH, NeupertC, Garcia-CantalejoJ, PopoloL, et al. (2011) The genetic interaction network of CCW12, a Saccharomyces cerevisiae gene required for cell wall integrity during budding and formation of mating projections. BMC Genomics 12 : 107.
57. YasharB, IrieK, PrintenJA, StevensonBJ, SpragueGFJr, et al. (1995) Yeast MEK-dependent signal transduction: response thresholds and parameters affecting fidelity. Mol Cell Biol 15 : 6545–6553.
58. Rodriguez-PenaJM, Diez-MunizS, BermejoC, NombelaC, ArroyoJ (2013) Activation of the yeast cell wall integrity MAPK pathway by zymolyase depends on protease and glucanase activities and requires the mucin-like protein Hkr1 but not Msb2. FEBS Lett 587 : 3675–3680.
59. AriasP, Diez-MunizS, GarciaR, NombelaC, Rodriguez-PenaJM, et al. (2011) Genome-wide survey of yeast mutations leading to activation of the yeast cell integrity MAPK pathway: novel insights into diverse MAPK outcomes. BMC Genomics 12 : 390.
60. CullenPJ, SpragueGFJr (2000) Glucose depletion causes haploid invasive growth in yeast. Proc Natl Acad Sci U S A 97 : 13619–13624.
61. CaustonHC, RenB, KohSS, HarbisonCT, KaninE, et al. (2001) Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell 12 : 323–337.
62. GaschAP, SpellmanPT, KaoCM, Carmel-HarelO, EisenMB, et al. (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11 : 4241–4257.
63. BicknellAA, TourtellotteJ, NiwaM (2010) Late phase of the endoplasmic reticulum stress response pathway is regulated by Hog1 MAP kinase. J Biol Chem 285 : 17545–17555.
64. Torres-QuirozF, Garcia-MarquesS, CoriaR, Randez-GilF, PrietoJA (2010) The activity of yeast Hog1 MAPK is required during endoplasmic reticulum stress induced by tunicamycin exposure. J Biol Chem 285 : 20088–20096.
65. NagalakshmiU, WangZ, WaernK, ShouC, RahaD, et al. (2008) The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320 : 1344–1349.
66. Randez-GilF, SanzP, EntianKD, PrietoJA (1998) Carbon source-dependent phosphorylation of hexokinase PII and its role in the glucose-signaling response in yeast. Mol Cell Biol 18 : 2940–2948.
67. HeifetzA, KeenanRW, ElbeinAD (1979) Mechanism of action of tunicamycin on the UDP-GlcNAc:dolichyl-phosphate Glc-NAc-1-phosphate transferase. Biochemistry 18 : 2186–2192.
68. BroC, KnudsenS, RegenbergB, OlssonL, NielsenJ (2005) Improvement of galactose uptake in Saccharomyces cerevisiae through overexpression of phosphoglucomutase: example of transcript analysis as a tool in inverse metabolic engineering. Appl Environ Microbiol 71 : 6465–6472.
69. LashkariDA, DeRisiJL, McCuskerJH, NamathAF, GentileC, et al. (1997) Yeast microarrays for genome wide parallel genetic and gene expression analysis. Proc Natl Acad Sci U S A 94 : 13057–13062.
70. TraversKJ, PatilCK, WodickaL, LockhartDJ, WeissmanJS, et al. (2000) Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101 : 249–258.
71. PatilC, WalterP (2001) Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr Opin Cell Biol 13 : 349–355.
72. AlepuzPM, JovanovicA, ReiserV, AmmererG (2001) Stress-induced map kinase Hog1 is part of transcription activation complexes. Mol Cell 7 : 767–777.
73. TatebayashiK, YamamotoK, TanakaK, TomidaT, MaruokaT, et al. (2006) Adaptor functions of Cdc42, Ste50, and Sho1 in the yeast osmoregulatory HOG MAPK pathway. Embo J 25 : 3033–3044.
74. KarunanithiS, CullenPJ (2012) The filamentous growth MAPK Pathway Responds to Glucose Starvation Through the Mig1/2 transcriptional repressors in Saccharomyces cerevisiae. Genetics 192 : 869–887.
75. SuranaU, RobitschH, PriceC, SchusterT, FitchI, et al. (1991) The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell 65 : 145–161.
76. BooherRN, DeshaiesRJ, KirschnerMW (1993) Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J 12 : 3417–3426.
77. ZahnerJE, HarkinsHA, PringleJR (1996) Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. Mol Cell Biol 16 : 1857–1870.
78. ChenT, HirokoT, ChaudhuriA, InoseF, LordM, et al. (2000) Multigenerational cortical inheritance of the Rax2 protein in orienting polarity and division in yeast. Science 290 : 1975–1978.
79. ChantJ, HerskowitzI (1991) Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell 65 : 1203–1212.
80. VernaJ, LodderA, LeeK, VagtsA, BallesterR (1997) A family of genes required for maintenance of cell wall integrity and for the stress response in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 94 : 13804–13809.
81. GavriasV, AndrianopoulosA, GimenoCJ, TimberlakeWE (1996) Saccharomyces cerevisiae TEC1 is required for pseudohyphal growth. Mol Microbiol 19 : 1255–1263.
82. MadhaniHD, FinkGR (1997) Combinatorial control required for the specificity of yeast MAPK signaling. Science 275 : 1314–1317.
83. HaoN, BeharM, ParnellSC, TorresMP, BorchersCH, et al. (2007) A systems-biology analysis of feedback inhibition in the Sho1 osmotic-stress-response pathway. Curr Biol 17 : 659–667.
84. BrewsterJL, de ValoirT, DwyerND, WinterE, GustinMC (1993) An osmosensing signal transduction pathway in yeast. Science 259 : 1760–1763.
85. MurakamiY, TatebayashiK, SaitoH (2008) Two adjacent docking sites in the yeast Hog1 mitogen-activated protein (MAP) kinase differentially interact with the Pbs2 MAP kinase kinase and the Ptp2 protein tyrosine phosphatase. Mol Cell Biol 28 : 2481–2494.
86. CarlsonM (1999) Glucose repression in yeast. Curr Opin Microbiol 2 : 202–207.
87. SchneperL, DuvelK, BroachJR (2004) Sense and sensibility: nutritional response and signal integration in yeast. Curr Opin Microbiol 7 : 624–630.
88. BhatPJ, MurthyTV (2001) Transcriptional control of the GAL/MEL regulon of yeast Saccharomyces cerevisiae: mechanism of galactose-mediated signal transduction. Mol Microbiol 40 : 1059–1066.
89. JohnstonM, FlickJS, PextonT (1994) Multiple mechanisms provide rapid and stringent glucose repression of GAL gene expression in Saccharomyces cerevisiae. Mol Cell Biol 14 : 3834–3841.
90. LohrD, VenkovP, ZlatanovaJ (1995) Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J 9 : 777–787.
91. JohnstonM (1987) A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev 51 : 458–476.
92. HoldenHM, RaymentI, ThodenJB (2003) Structure and function of enzymes of the Leloir pathway for galactose metabolism. J Biol Chem 278 : 43885–43888.
93. Melcher K (1997) Galactose metabolism in Saccharomyces cerevisiae: a paradigm for eukaryotic gene regulation. FK Zimmermann, K-D Entian (Eds), Yeast Sugar Metabolism, Technomic Publishing Inc, Lancaster, PA 235–269.
94. GruningNM, RinnerthalerM, BluemleinK, MullederM, WamelinkMM, et al. (2011) Pyruvate kinase triggers a metabolic feedback loop that controls redox metabolism in respiring cells. Cell Metab 14 : 415–427.
95. RuckenstuhlC, ButtnerS, Carmona-GutierrezD, EisenbergT, KroemerG, et al. (2009) The Warburg effect suppresses oxidative stress induced apoptosis in a yeast model for cancer. PLoS One 4: e4592.
96. AlexandreA, LehningerAL (1984) Bypasses of the antimycin a block of mitochondrial electron transport in relation to ubisemiquinone function. Biochim Biophys Acta 767 : 120–129.
97. PhamNA, RobinsonBH, HedleyDW (2000) Simultaneous detection of mitochondrial respiratory chain activity and reactive oxygen in digitonin-permeabilized cells using flow cytometry. Cytometry 41 : 245–251.
98. CampoML, KinnallyKW, TedeschiH (1992) The effect of antimycin A on mouse liver inner mitochondrial membrane channel activity. J Biol Chem 267 : 8123–8127.
99. CelenzaJL, CarlsonM (1986) A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science 233 : 1175–1180.
100. SchullerHJ, EntianKD (1991) Extragenic suppressors of yeast glucose derepression mutants leading to constitutive synthesis of several glucose-repressible enzymes. J Bacteriol 173 : 2045–2052.
101. McCartneyRR, SchmidtMC (2001) Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J Biol Chem 276 : 36460–36466.
102. TreitelMA, KuchinS, CarlsonM (1998) Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol Cell Biol 18 : 6273–6280.
103. OstlingJ, RonneH (1998) Negative control of the Mig1p repressor by Snf1p-dependent phosphorylation in the absence of glucose. Eur J Biochem 252 : 162–168.
104. SmithFC, DaviesSP, WilsonWA, CarlingD, HardieDG (1999) The SNF1 kinase complex from Saccharomyces cerevisiae phosphorylates the transcriptional repressor protein Mig1p in vitro at four sites within or near regulatory domain 1. FEBS Lett 453 : 219–223.
105. ZhouH, WinstonF (2001) NRG1 is required for glucose repression of the SUC2 and GAL genes of Saccharomyces cerevisiae. Bmc Genetics 2 : 5.
106. VallierLG, CarlsonM (1994) Synergistic release from glucose repression by mig1 and ssn mutations in Saccharomyces cerevisiae. Genetics 137 : 49–54.
107. OrlovaM, OzcetinH, BarrettL, KuchinS (2010) Roles of the Snf1-activating kinases during nitrogen limitation and pseudohyphal differentiation in Saccharomyces cerevisiae. Eukaryot Cell 9 : 208–214.
108. HerscovicsA, OrleanP (1993) Glycoprotein biosynthesis in yeast. FASEB J 7 : 540–550.
109. GracyRW, NoltmannEA (1968) Studies on phosphomannose isomerase. II. Characterization as a zinc metalloenzyme. J Biol Chem 243 : 4109–4116.
110. SmithDJ, ProudfootA, FriedliL, KligLS, ParaviciniG, et al. (1992) PMI40, an intron-containing gene required for early steps in yeast mannosylation. Mol Cell Biol 12 : 2924–2930.
111. CoxJS, ShamuCE (1993) WalterP (1993) Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73 : 1197–1206.
112. RonD, WalterP (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol 8 : 519–529.
113. CoxJS, WalterP (1996) A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87 : 391–404.
114. TanigawaM, KiharaA, TerashimaM, TakaharaT, MaedaT (2012) Sphingolipids regulate the yeast high-osmolarity glycerol response pathway. Mol Cell Biol 32 : 2861–2870.
115. SinghKK (2000) The Saccharomyces cerevisiae Sln1p-Ssk1p two-component system mediates response to oxidative stress and in an oxidant-specific fashion. Free Radic Biol Med 29 : 1043–1050.
116. HaghnazariE, HeyerWD (2004) The Hog1 MAP kinase pathway and the Mec1 DNA damage checkpoint pathway independently control the cellular responses to hydrogen peroxide. DNA Repair (Amst) 3 : 769–776.
117. Wurgler-MurphySM, MaedaT, WittenEA, SaitoH (1997) Regulation of the Saccharomyces cerevisiae HOG1 mitogen-activated protein kinase by the PTP2 and PTP3 protein tyrosine phosphatases. Mol Cell Biol 17 : 1289–1297.
118. LongtineMS, DeMariniDJ, ValencikML, Al-AwarOS, FaresH, et al. (1996) The septins: roles in cytokinesis and other processes. Curr Opin Cell Biol 8 : 106–119.
119. GladfelterAS, PringleJR, LewDJ (2001) The septin cortex at the yeast mother-bud neck. Curr Opin Microbiol 4 : 681–689.
120. ReynoldsTB, FinkGR (2001) Bakers' yeast, a model for fungal biofilm formation. Science 291 : 878–881.
121. KarunanithiS, JoshiJ, ChavelC, BirkayaB, GrellL, et al. (2012) Regulation of mat responses by a differentiation MAPK pathway in Saccharomyces cerevisiae. PLoS ONE 7: e32294.
122. KarunanithiS, VadaieN, ChavelCA, BirkayaB, JoshiJ, et al. (2010) Shedding of the mucin-like flocculin Flo11p reveals a new aspect of fungal adhesion regulation. Curr Biol 20 : 1389–1395.
123. ChenJ, ChenJ, LaneS, LiuH (2002) A conserved mitogen-activated protein kinase pathway is required for mating in Candida albicans. Mol Microbiol 46 : 1335–1344.
124. CsankC, SchroppelK, LebererE, HarcusD, MohamedO, et al. (1998) Roles of the Candida albicans mitogen-activated protein kinase homolog, Cek1p, in hyphal development and systemic candidiasis. Infect Immun 66 : 2713–2721.
125. KohlerJR, FinkGR (1996) Candida albicans strains heterozygous and homozygous for mutations in mitogen-activated protein kinase signaling components have defects in hyphal development. Proc Natl Acad Sci U S A 93 : 13223–13228.
126. San JoseC, MongeRA, Perez-DiazR, PlaJ, NombelaC (1996) The mitogen-activated protein kinase homolog HOG1 gene controls glycerol accumulation in the pathogenic fungus Candida albicans. J Bacteriol 178 : 5850–5852.
127. SmithDA, NichollsS, MorganBA, BrownAJ, QuinnJ (2004) A conserved stress-activated protein kinase regulates a core stress response in the human pathogen Candida albicans. Mol Biol Cell 15 : 4179–4190.
128. EismanB, Alonso-MongeR, RomanE, AranaD, NombelaC, et al. (2006) The Cek1 and Hog1 mitogen-activated protein kinases play complementary roles in cell wall biogenesis and chlamydospore formation in the fungal pathogen Candida albicans. Eukaryot Cell 5 : 347–358.
129. MitchellAP (1998) Dimorphism and virulence in Candida albicans. Curr Opin Microbiol 1 : 687–692.
130. BiswasS, RoyM, DattaA (2003) N-acetylglucosamine-inducible CaGAP1 encodes a general amino acid permease which co-ordinates external nitrogen source response and morphogenesis in Candida albicans. Microbiology 149 : 2597–2608.
131. LawrenceCL, BottingCH, AntrobusR, CootePJ (2004) Evidence of a new role for the high-osmolarity glycerol mitogen-activated protein kinase pathway in yeast: regulating adaptation to citric acid stress. Mol Cell Biol 24 : 3307–3323.
132. HickmanMJ, SpattD, WinstonF (2011) The Hog1 mitogen-activated protein kinase mediates a hypoxic response in Saccharomyces cerevisiae. Genetics 188 : 325–338.
133. PanaderoJ, PallottiC, Rodriguez-VargasS, Randez-GilF, PrietoJA (2006) A downshift in temperature activates the high osmolarity glycerol (HOG) pathway, which determines freeze tolerance in Saccharomyces cerevisiae. J Biol Chem 281 : 4638–4645.
134. VendrellA, PosasF (2011) Sir2 plays a key role in cell fate determination upon SAPK activation. Aging (Albany NY) 3 : 1163–1168.
135. FanM, RheeJ, St-PierreJ, HandschinC, PuigserverP, et al. (2004) Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1alpha: modulation by p38 MAPK. Genes Dev 18 : 278–289.
136. PuigserverP, RheeJ, LinJ, WuZ, YoonJC, et al. (2001) Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARgamma coactivator-1. Mol Cell 8 : 971–982.
137. XiX, HanJ, ZhangJZ (2001) Stimulation of glucose transport by AMP-activated protein kinase via activation of p38 mitogen-activated protein kinase. J Biol Chem 276 : 41029–41034.
138. LinCC, ChengTL, TsaiWH, TsaiHJ, HuKH, et al. (2012) Loss of the respiratory enzyme citrate synthase directly links the Warburg effect to tumor malignancy. Sci Rep 2 : 785.
139. CoxJS, ChapmanRE, WalterP (1997) The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol Biol Cell 8 : 1805–1814.
140. MearesGP, HughesKJ, NaatzA, PapaFR, UranoF, et al. (2011) IRE1-dependent activation of AMPK in response to nitric oxide. Mol Cell Biol 31 : 4286–4297.
141. KomurovK, TsengJT, MullerM, SeviourEG, MossTJ, et al. (2012) The glucose-deprivation network counteracts lapatinib-induced toxicity in resistant ErbB2-positive breast cancer cells. Mol Syst Biol 8 : 596.
142. ChenX, IliopoulosD, ZhangQ, TangQ, GreenblattMB, et al. (2014) XBP1 promotes triple-negative breast cancer by controlling the HIF1alpha pathway. Nature 508 : 103–107.
143. YoungRM, AckermanD, QuinnZL, MancusoA, GruberM, et al. (2013) Dysregulated mTORC1 renders cells critically dependent on desaturated lipids for survival under tumor-like stress. Genes Dev 27 : 1115–1131.
144. GreenblattMB, ShimJH, GlimcherLH (2013) Mitogen-activated protein kinase pathways in osteoblasts. Annu Rev Cell Dev Biol 29 : 63–79.
145. TrovatiM, DoronzoG, BaraleC, VaccherisC, RussoI, et al. (2014) Leptin and vascular smooth muscle cells. Curr Pharm Des 20 : 625–634.
146. Sambrook J FEF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
147. Rose MD, Winston F., and Hieter P. (1990) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
148. RobertsRL, FinkGR (1994) Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev 8 : 2974–2985.
149. FaresH, GoetschL, PringleJR (1996) Identification of a developmentally regulated septin and involvement of the septins in spore formation in Saccharomyces cerevisiae. J Cell Biol 132 : 399–411.
150. MadhaniHD, StylesCA, FinkGR (1997) MAP kinases with distinct inhibitory functions impart signaling specificity during yeast differentiation. Cell 91 : 673–684.
151. EllisCD, WangF, MacDiarmidCW, ClarkS, LyonsT, et al. (2004) Zinc and the Msc2 zinc transporter protein are required for endoplasmic reticulum function. J Cell Biol 166 : 325–335.
152. GoldsteinAL, McCuskerJH (1999) Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15 : 1541–1553.
153. BaudinA, Ozier-KalogeropoulosO, DenouelA, LacrouteF, CullinC (1993) A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res 21 : 3329–3330.
154. TrapnellC, PachterL, SalzbergSL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25 : 1105–1111.
155. RobinsonMD, McCarthyDJ, SmythGK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26 : 139–140.
156. ReinerA, YekutieliD, BenjaminiY (2003) Identifying differentially expressed genes using false discovery rate controlling procedures. Bioinformatics 19 : 368–375.
157. LivakKJ, SchmittgenTD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 25 : 402–408.
158. LeeMJ, DohlmanHG (2008) Coactivation of G protein signaling by cell-surface receptors and an intracellular exchange factor. Curr Biol 18 : 211–215.
159. SchneiderCA, RasbandWS, EliceiriKW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9 : 671–675.
160. RyanO, ShapiroRS, KuratCF, MayhewD, BaryshnikovaA, et al. (2012) Global gene deletion analysis exploring yeast filamentous growth. Science 337 : 1353–1356.
161. LiuH, StylesCA, FinkGR (1993) Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science 262 : 1741–1744.
162. FonziWA, IrwinMY (1993) Isogenic strain construction and gene mapping in Candida albicans. Genetics 134 : 717–728.
163. BlankenshipJR, FanningS, HamakerJJ, MitchellAP (2010) An extensive circuitry for cell wall regulation in Candida albicans. PLoS Pathog 6: e1000752.
Štítky
Genetika Reprodukčná medicína
Článek Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 LociČlánek The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in MeiosisČlánek Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective PressuresČlánek Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome ReductionČlánek BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian SkinČlánek Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in ArabidopsisČlánek RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress ResponseČlánek Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping StudiesČlánek Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation inČlánek Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) SeedlingsČlánek Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition inČlánek A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) DegradationČlánek Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2014 Číslo 10- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- An Deletion Is Highly Associated with a Juvenile-Onset Inherited Polyneuropathy in Leonberger and Saint Bernard Dogs
- Licensing of Yeast Centrosome Duplication Requires Phosphoregulation of Sfi1
- Oligoasthenoteratozoospermia and Infertility in Mice Deficient for miR-34b/c and miR-449 Loci
- Basement Membrane and Cell Integrity of Self-Tissues in Maintaining Immunological Tolerance
- The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in Meiosis
- Germline Mutations in Are Associated with Familial Gastric Cancer
- POT1a and Components of CST Engage Telomerase and Regulate Its Activity in
- Controlling Meiotic Recombinational Repair – Specifying the Roles of ZMMs, Sgs1 and Mus81/Mms4 in Crossover Formation
- Payoffs, Not Tradeoffs, in the Adaptation of a Virus to Ostensibly Conflicting Selective Pressures
- FHIT Suppresses Epithelial-Mesenchymal Transition (EMT) and Metastasis in Lung Cancer through Modulation of MicroRNAs
- Genome-Wide Mapping of Yeast RNA Polymerase II Termination
- Examination of Prokaryotic Multipartite Genome Evolution through Experimental Genome Reduction
- White Cells Facilitate Opposite- and Same-Sex Mating of Opaque Cells in
- BMP-FGF Signaling Axis Mediates Wnt-Induced Epidermal Stratification in Developing Mammalian Skin
- Genome-Wide Association Study of CSF Levels of 59 Alzheimer's Disease Candidate Proteins: Significant Associations with Proteins Involved in Amyloid Processing and Inflammation
- COE Loss-of-Function Analysis Reveals a Genetic Program Underlying Maintenance and Regeneration of the Nervous System in Planarians
- Fat-Dachsous Signaling Coordinates Cartilage Differentiation and Polarity during Craniofacial Development
- Identification of Genes Important for Cutaneous Function Revealed by a Large Scale Reverse Genetic Screen in the Mouse
- Sensors at Centrosomes Reveal Determinants of Local Separase Activity
- Genes Integrate and Hedgehog Pathways in the Second Heart Field for Cardiac Septation
- Systematic Dissection of Coding Exons at Single Nucleotide Resolution Supports an Additional Role in Cell-Specific Transcriptional Regulation
- Recovery from an Acute Infection in Requires the GATA Transcription Factor ELT-2
- HIPPO Pathway Members Restrict SOX2 to the Inner Cell Mass Where It Promotes ICM Fates in the Mouse Blastocyst
- Role of and in Development of Abdominal Epithelia Breaks Posterior Prevalence Rule
- The Formation of Endoderm-Derived Taste Sensory Organs Requires a -Dependent Expansion of Embryonic Taste Bud Progenitor Cells
- Role of STN1 and DNA Polymerase α in Telomere Stability and Genome-Wide Replication in Arabidopsis
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- Encodes the Catalytic Subunit of N Alpha-Acetyltransferase that Regulates Development, Metabolism and Adult Lifespan
- Disruption of SUMO-Specific Protease 2 Induces Mitochondria Mediated Neurodegeneration
- Caudal Regulates the Spatiotemporal Dynamics of Pair-Rule Waves in
- It's All in Your Mind: Determining Germ Cell Fate by Neuronal IRE-1 in
- A Conserved Role for Homologs in Protecting Dopaminergic Neurons from Oxidative Stress
- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- An AGEF-1/Arf GTPase/AP-1 Ensemble Antagonizes LET-23 EGFR Basolateral Localization and Signaling during Vulva Induction
- The Proteomic Landscape of the Suprachiasmatic Nucleus Clock Reveals Large-Scale Coordination of Key Biological Processes
- RNA-Processing Protein TDP-43 Regulates FOXO-Dependent Protein Quality Control in Stress Response
- A Complex Genetic Switch Involving Overlapping Divergent Promoters and DNA Looping Regulates Expression of Conjugation Genes of a Gram-positive Plasmid
- ZTF-8 Interacts with the 9-1-1 Complex and Is Required for DNA Damage Response and Double-Strand Break Repair in the Germline
- Integrating Functional Data to Prioritize Causal Variants in Statistical Fine-Mapping Studies
- Tpz1-Ccq1 and Tpz1-Poz1 Interactions within Fission Yeast Shelterin Modulate Ccq1 Thr93 Phosphorylation and Telomerase Recruitment
- Salt-Induced Stabilization of EIN3/EIL1 Confers Salinity Tolerance by Deterring ROS Accumulation in
- Telomeric (s) in spp. Encode Mediator Subunits That Regulate Distinct Virulence Traits
- Ethylene-Induced Inhibition of Root Growth Requires Abscisic Acid Function in Rice ( L.) Seedlings
- Ancient Expansion of the Hox Cluster in Lepidoptera Generated Four Homeobox Genes Implicated in Extra-Embryonic Tissue Formation
- Mechanism of Suppression of Chromosomal Instability by DNA Polymerase POLQ
- A Mutation in the Mouse Gene Leads to Impaired Hedgehog Signaling
- Keeping mtDNA in Shape between Generations
- Targeted Exon Capture and Sequencing in Sporadic Amyotrophic Lateral Sclerosis
- TIF-IA-Dependent Regulation of Ribosome Synthesis in Muscle Is Required to Maintain Systemic Insulin Signaling and Larval Growth
- At Short Telomeres Tel1 Directs Early Replication and Phosphorylates Rif1
- Evidence of a Bacterial Receptor for Lysozyme: Binding of Lysozyme to the Anti-σ Factor RsiV Controls Activation of the ECF σ Factor σ
- Hsp40s Specify Functions of Hsp104 and Hsp90 Protein Chaperone Machines
- Feeding State, Insulin and NPR-1 Modulate Chemoreceptor Gene Expression via Integration of Sensory and Circuit Inputs
- Functional Interaction between Ribosomal Protein L6 and RbgA during Ribosome Assembly
- Multiple Regulatory Systems Coordinate DNA Replication with Cell Growth in
- Fast Evolution from Precast Bricks: Genomics of Young Freshwater Populations of Threespine Stickleback
- Mmp1 Processing of the PDF Neuropeptide Regulates Circadian Structural Plasticity of Pacemaker Neurons
- The Nuclear Immune Receptor Is Required for -Dependent Constitutive Defense Activation in
- Genetic Modifiers of Neurofibromatosis Type 1-Associated Café-au-Lait Macule Count Identified Using Multi-platform Analysis
- Juvenile Hormone-Receptor Complex Acts on and to Promote Polyploidy and Vitellogenesis in the Migratory Locust
- Uncovering Enhancer Functions Using the α-Globin Locus
- The Analysis of Mutant Alleles of Different Strength Reveals Multiple Functions of Topoisomerase 2 in Regulation of Chromosome Structure
- Metabolic Respiration Induces AMPK- and Ire1p-Dependent Activation of the p38-Type HOG MAPK Pathway
- The Specification and Global Reprogramming of Histone Epigenetic Marks during Gamete Formation and Early Embryo Development in
- The DAF-16 FOXO Transcription Factor Regulates to Modulate Stress Resistance in , Linking Insulin/IGF-1 Signaling to Protein N-Terminal Acetylation
- Genetic Influences on Translation in Yeast
- Analysis of Mutants Defective in the Cdk8 Module of Mediator Reveal Links between Metabolism and Biofilm Formation
- Ribosomal Readthrough at a Short UGA Stop Codon Context Triggers Dual Localization of Metabolic Enzymes in Fungi and Animals
- Gene Duplication Restores the Viability of Δ and Δ Mutants
- Selection on a Variant Associated with Improved Viral Clearance Drives Local, Adaptive Pseudogenization of Interferon Lambda 4 ()
- Break-Induced Replication Requires DNA Damage-Induced Phosphorylation of Pif1 and Leads to Telomere Lengthening
- Dynamic Partnership between TFIIH, PGC-1α and SIRT1 Is Impaired in Trichothiodystrophy
- Signature Gene Expression Reveals Novel Clues to the Molecular Mechanisms of Dimorphic Transition in
- Mutations in Moderate or Severe Intellectual Disability
- Multifaceted Genome Control by Set1 Dependent and Independent of H3K4 Methylation and the Set1C/COMPASS Complex
- A Role for Taiman in Insect Metamorphosis
- The Small RNA Rli27 Regulates a Cell Wall Protein inside Eukaryotic Cells by Targeting a Long 5′-UTR Variant
- MMS Exposure Promotes Increased MtDNA Mutagenesis in the Presence of Replication-Defective Disease-Associated DNA Polymerase γ Variants
- Coexistence and Within-Host Evolution of Diversified Lineages of Hypermutable in Long-term Cystic Fibrosis Infections
- Comprehensive Mapping of the Flagellar Regulatory Network
- Topoisomerase II Is Required for the Proper Separation of Heterochromatic Regions during Female Meiosis
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- KDM5 Interacts with Foxo to Modulate Cellular Levels of Oxidative Stress
- H2B Mono-ubiquitylation Facilitates Fork Stalling and Recovery during Replication Stress by Coordinating Rad53 Activation and Chromatin Assembly
- Copy Number Variation in the Horse Genome
- Unifying Genetic Canalization, Genetic Constraint, and Genotype-by-Environment Interaction: QTL by Genomic Background by Environment Interaction of Flowering Time in
- Spinster Homolog 2 () Deficiency Causes Early Onset Progressive Hearing Loss
- Genome-Wide Discovery of Drug-Dependent Human Liver Regulatory Elements
- Developmentally-Regulated Excision of the SPβ Prophage Reconstitutes a Gene Required for Spore Envelope Maturation in
- Protein Phosphatase 4 Promotes Chromosome Pairing and Synapsis, and Contributes to Maintaining Crossover Competence with Increasing Age
- The bHLH-PAS Transcription Factor Dysfusion Regulates Tarsal Joint Formation in Response to Notch Activity during Leg Development
- A Mouse Model Uncovers LKB1 as an UVB-Induced DNA Damage Sensor Mediating CDKN1A (p21) Degradation
- Notch3 Interactome Analysis Identified WWP2 as a Negative Regulator of Notch3 Signaling in Ovarian Cancer
- An Integrated Cell Purification and Genomics Strategy Reveals Multiple Regulators of Pancreas Development
- Dominant Sequences of Human Major Histocompatibility Complex Conserved Extended Haplotypes from to
- The Vesicle Protein SAM-4 Regulates the Processivity of Synaptic Vesicle Transport
- A Gain-of-Function Mutation in Impeded Bone Development through Increasing Expression in DA2B Mice
- Nephronophthisis-Associated Regulates Cell Cycle Progression, Apoptosis and Epithelial-to-Mesenchymal Transition
- Beclin 1 Is Required for Neuron Viability and Regulates Endosome Pathways via the UVRAG-VPS34 Complex
- The Not5 Subunit of the Ccr4-Not Complex Connects Transcription and Translation
- Abnormal Dosage of Ultraconserved Elements Is Highly Disfavored in Healthy Cells but Not Cancer Cells
- Genome-Wide Distribution of RNA-DNA Hybrids Identifies RNase H Targets in tRNA Genes, Retrotransposons and Mitochondria
- The Chromosomal Association of the Smc5/6 Complex Depends on Cohesion and Predicts the Level of Sister Chromatid Entanglement
- Cell-Autonomous Progeroid Changes in Conditional Mouse Models for Repair Endonuclease XPG Deficiency
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- The Master Activator of IncA/C Conjugative Plasmids Stimulates Genomic Islands and Multidrug Resistance Dissemination
- A Splice Mutation in the Gene Causes High Glycogen Content and Low Meat Quality in Pig Skeletal Muscle
- Keratin 76 Is Required for Tight Junction Function and Maintenance of the Skin Barrier
- A Role for Taiman in Insect Metamorphosis
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



