-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
Recurrent Evolution of Melanism in South American Felids
Color polymorphism in closely related animal species provides an opportunity to study how the balance between natural selection and genetic drift shapes the evolution of appearance and form. The cat family, Felidae, is especially interesting; 13 of 37 extant species exhibit polymorphism for melanism, but evidence for any adaptive role is lacking, in part because the potential benefits of melanism to felid predators are not clear, and in part because the tools for genomic analysis of natural populations are limited. We identify the mutations responsible for melanism in three closely related South American wild felids, the pampas cat, the kodkod, and Geoffroy’s cat, then adapt a new approach for targeted genome sequencing to characterize molecular variation in the region surrounding each melanism mutation. We find that each mutation has developed independently, with strong evidence for natural selection in the black pampas cat, and reduced genetic variation in the entire population of kodkods. Our results demonstrate that some “black cats” are black not by chance, but by selection for a mutation that provides increased fitness.
Published in the journal: Recurrent Evolution of Melanism in South American Felids. PLoS Genet 11(2): e32767. doi:10.1371/journal.pgen.1004892
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004892Summary
Color polymorphism in closely related animal species provides an opportunity to study how the balance between natural selection and genetic drift shapes the evolution of appearance and form. The cat family, Felidae, is especially interesting; 13 of 37 extant species exhibit polymorphism for melanism, but evidence for any adaptive role is lacking, in part because the potential benefits of melanism to felid predators are not clear, and in part because the tools for genomic analysis of natural populations are limited. We identify the mutations responsible for melanism in three closely related South American wild felids, the pampas cat, the kodkod, and Geoffroy’s cat, then adapt a new approach for targeted genome sequencing to characterize molecular variation in the region surrounding each melanism mutation. We find that each mutation has developed independently, with strong evidence for natural selection in the black pampas cat, and reduced genetic variation in the entire population of kodkods. Our results demonstrate that some “black cats” are black not by chance, but by selection for a mutation that provides increased fitness.
Introduction
Color variation in natural populations is a useful entry point to investigate the evolution of mammalian phenotypic diversity. Pigmentary differences can be readily observed and quantified; much is known about the underlying biochemistry and cell biology, and there are many examples of apparent convergent evolution. Fundamental questions such as the relative roles played by regulatory vs. protein-coding variation, the phylogenetic origin of similar phenotypes shared among different lineages, and the potential impact of pleiotropic mutations can all be explored from a molecular genetic perspective [1–3].
Mammalian color diversity is particularly apparent in the Felidae, with a wide range of patterns and base colors represented among 37 species with a common ancestor ~11 million years ago [4,5]. In addition to color patterns such as rosettes, stripes, or spots, two characteristic pigmentary phenotypes appear in multiple species [6,7]. The ticked phenotype, a brushed appearance that hides dark tabby markings, is characteristic of 4 wild species (the lion, puma, caracal, and jaguarundi), and melanism, a black coat with residual or “ghost” dark tabby markings in some individuals, has been described in 13 wild species (see [8] for references). Felid melanism is especially interesting because it is polymorphic within each species, but its possible adaptive significance has received little attention to date [9]. In rodents and in birds, an adaptive role of melanism is well-established from ecological and field studies [1,2,10,11], but the evolutionary forces that underlie melanism in larger mammals are less clear, with examples of introgression in the case of North American black wolves [12], and a founder effect in the case of Malaysian black leopards [13]. A recent study by Allen et al. [9] found that the presence of melanism across felid species is correlated with habitat and/or behavioral diversity, and suggested a common underlying mechanism of disruptive selection.
A particularly interesting group of animals from this perspective is an endemic lineage of Neotropical wild cats that includes multiple species exhibiting melanism as a naturally occurring phenotypic variant. This lineage comprises eight species of small cats belonging to the genus Leopardus, which diverged from each other after the colonization of South America by a common ancestor ~3 million years ago (MYA) [4,5]. In three of these species, the pampas cat (L. colocolo), the kodkod (L. guigna, referred to locally as the güiña or hüiña in Chile or Argentina, respectively), and Geoffroy’s cat (L. geoffroyi), melanistic individuals comprise 20% or more of the population in some areas [14–17]. Inter-species hybridization within Leopardus has been reported [18,19]; thus, high frequencies of melanism within these species could reflect introgression, an ancient trans-specific polymorphism, or independent evolution after divergence from a common ancestor.
To distinguish among these possibilities and to better understand how similar phenotypes evolve in closely related felids, we used massively parallel targeted resequencing to delineate the molecular cause and population genetic history of melanism mutations in each of the three species. The closest reference genome to Leopardus spp. is that of the domestic cat, with a last common ancestor ~6 MYA. As an alternative to oligonucleotide capture hybridization, which is constrained to protein-coding regions of closely related species [20], we adapted an approach in which large insert clones (from bacterial artificial chromosome or fosmid vectors) are used as a template to generate biotinylated RNA hybridization probes for solution hybridization capture of homologous segments in the three species. This approach, termed CATCH-Seq (for “Clone Adapted Template Capture Hybridization”) was developed to assess dense human genetic variation in regions that are otherwise difficult to assess by other methods of genotyping [21], e.g. the MHC, but, as shown here, can be extended to facilitate resequencing of targeted genomic regions in any organism for which clone-based reagents from a related species are available.
We identify different molecular causes of melanism in the pampas cat, the kodkod, and the Geoffroy’s cat. Haplotype-based analyses point to distinctive population histories in all three species, with one—the pampas cat—exhibiting strong evidence of selection for melanism. Our results have implications for pigmentary biology and genetics, and yield new insight into the evolution of wild felids.
Results
Populations and samples
The pampas and Geoffroy’s cats are widely distributed through South America, while the kodkod is geographically more restricted (Fig. 1). All three species, which diverged from a common ancestor ~2.5 MYA, are relatively small, inhabit savanna and grassland areas as well as temperate rainforests (particularly the kodkod), and prey on small mammals, birds, and/or amphibians and reptiles. To mitigate against confounding effects of population structure, we sampled individuals from well-defined geographic regions (Fig. 1) in Emas National Park (Brazil), the southern region of Rio Grande do Sul (Brazil), and Chiloé Island (Chile), where the frequencies of melanism are 25%, 20%, and 29% for the pampas cat, Geoffroy’s cat and kodkod, respectively (Table 1, Table S1 in S1 Data).
Fig. 1. Melanism mutations and phenotypes in three Leopardus species 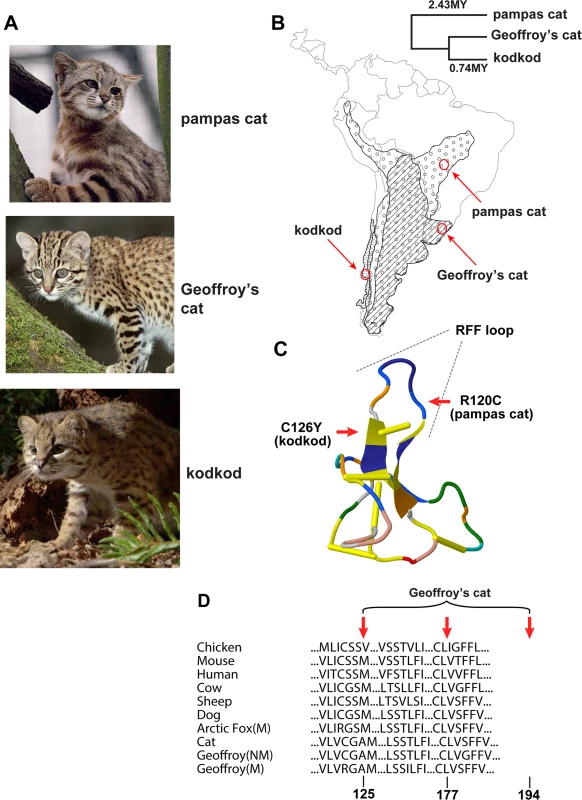
(A) Color phenotypes of the pampas cat (L. colocolo), Geoffroy’s cat (L. geoffroyi), and kodkod (L. guigna). Tabby markings in the non-melanistic (NM) forms shown here are obscured in the melanistic (M) forms. (B) Geographic distribution (modified from [7]); red circles represent the approximate origin of samples as described in the text. Right upper panel indicates phylogenetic relationships and estimated divergence times based on [4] (C, D) Location of missense variants in ASIP (kodkod, pampas cat) and MC1R (Geoffroy’s cat). (C) The kodkod and pampas cat variants are predicted to disrupt the three dimensional structure of a disulfide-stabilized loop in ASIP that is critical for MC1R binding [30]. (D) The Geoffroy’s cat melanism allele carries three missense variants in MC1R; C125R is likely to be causative as described in the text. Tab. 1. Genotype, allele, and phenotype frequencies for melanism variants. 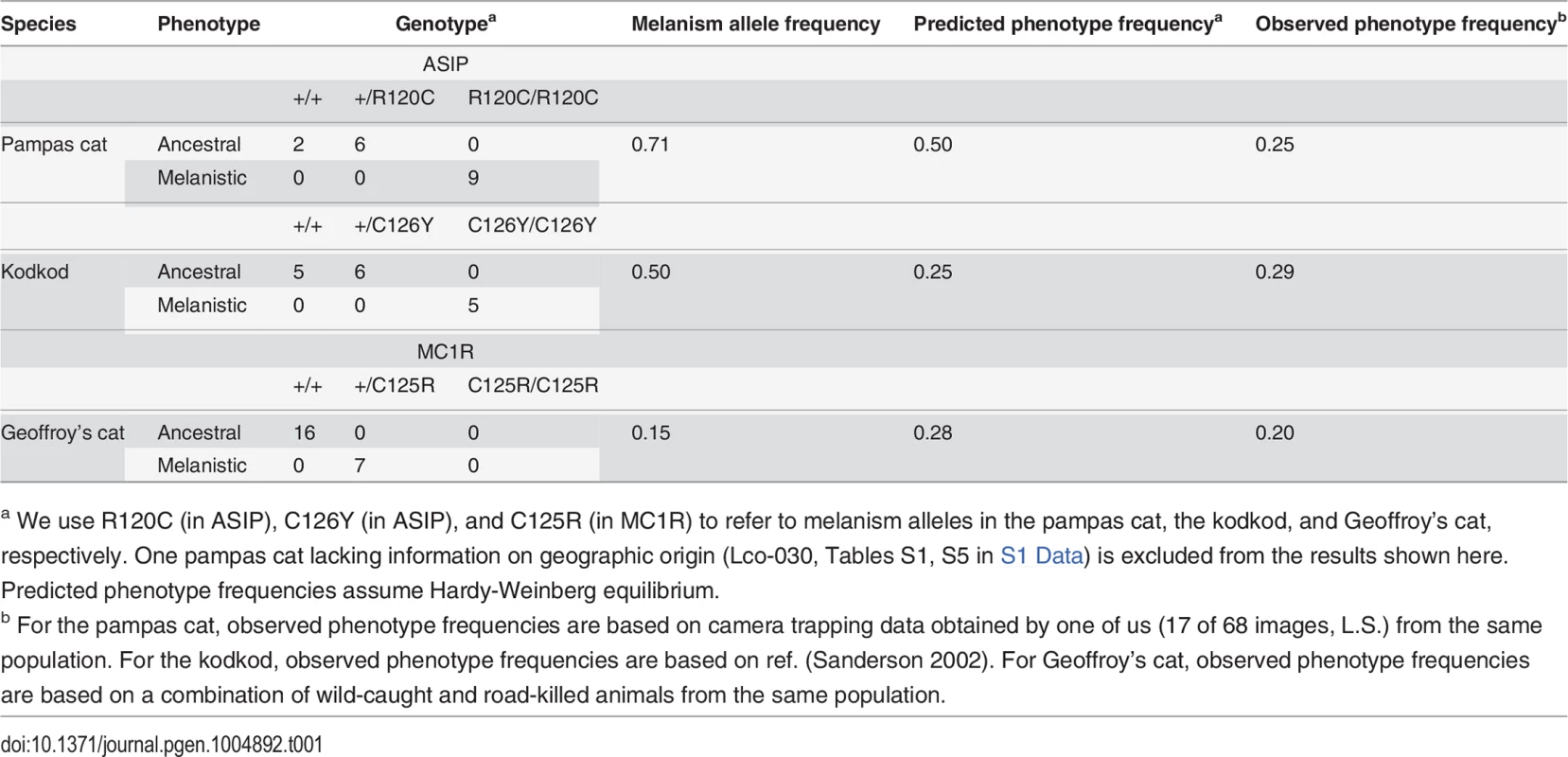
a We use R120C (in ASIP), C126Y (in ASIP), and C125R (in MC1R) to refer to melanism alleles in the pampas cat, the kodkod, and Geoffroy’s cat, respectively. One pampas cat lacking information on geographic origin (Lco-030, Tables S1, S5 in S1 Data) is excluded from the results shown here. Predicted phenotype frequencies assume Hardy-Weinberg equilibrium. Melanism and pigment type-switching: mutations in Agouti signaling protein (ASIP) and Melanocortin 1 receptor (MC1R)
CATCH-Seq is best suited to explore sequence variation once specific regions of interest are identified; therefore we initiated molecular analyses of melanism by conventional capillary-based sequencing of candidate genes. In mammals, melanism represents a shift in the balance between red-yellow pheomelanin and brown-black eumelanin, controlled by the activity of a melanocyte-specific G protein-coupled receptor, the Melanocortin 1 receptor (MC1R) [22–25]. The most common causes of melanism mutations are gain-of-function alterations in MC1R, or loss-of-function alterations in ASIP, which encodes Agouti signaling protein, a paracrine signaling molecule that inhibits MC1R signaling [26]. Consistent with the nature of the mutations, melanism caused by MC1R mutations is dominantly inherited, while melanism caused by ASIP mutations is recessively inherited; however, the inheritance pattern of melanism in pampas, Geoffroy’s cat, and the kodkod is not known. (Additional, less frequent causes of melanism in some mammals include mutations of beta-defensin 103 [an alternative ligand for MC1R] [27], Attractin [an accessory receptor for ASIP] [28], or Mahogunin [an E3 ubiquitin ligase that acts upstream of the MC1R] [29]).
Using the domestic cat genome as a starting point for the design of oligonucleotide PCR primers, we determined the protein-coding sequence of ASIP and MC1R in 18 pampas cats (10 melanistic), 23 Geoffroy’s cats (7 melanistic), and 16 kodkods (5 melanistic). Animals were selected from specific geographic areas as described above, and collection was independent of coat color phenotype (Table S1 in S1 Data). In each species, we identified missense alterations whose molecular nature, pattern of association, and apparent mode of inheritance made a compelling case for them being the cause of melanism (Table 1, Table S4 in S1 Data, Fig. 1).
In both the pampas cat and the kodkod, mutations in ASIP were identified that are predicted to cause a loss-of-function, and homozygosity for these mutations was completely associated with melanism. In the pampas cat, an inferred Arg to Cys substitution in the C-terminal region of ASIP (p.R120C) lies in the critical RFF loop (e.g. a triplet motif made of one arginine and two phenylalanine residues), required for binding to the MC1R [30], and also creates an odd number of cysteine residues, predicted to interfere with folding of the disulfide-rich domain. Similarly, in the kodkod, an inferred Cys to Tyr substitution in ASIP (p.C126Y) affects the key disulfide bond that stabilizes the RFF loop [30]. All melanistic pampas cat individuals were homozygous for ASIPR120C (Chi-square = 18, p<0.005) and all melanistic kodkod indivduals were homozygous for ASIPC126Y (Chi-square = 16, p<0.005). No other ASIP coding sequence variants were identified in the pampas cat or in the kodkod (Table S5 in S1 Data); four intraspecific variants in MC1R were present in the pampas cat, but none of them exhibited an association with melanism (Table S4 in S1 Data).
In Geoffroy’s cat, we identified an MC1R mutation as the likely cause of melanism. All melanistic individuals were heterozygous for four variants that predicted three nonsynonymous substitutions, p.C125R, p.T177I, and p.G194S (Table S6 in S1 Data). All individuals were either homozygous for the ancestral alleles (125Cys, 177Thr, 194Gly) or heterozygous for the derivative alleles (125Arg, 177Ile, 194Ser); we observed complete association of heterozygosity for the derivative alleles with melanism (Chi-square = 16, p<0.005). In the laboratory mouse, a Cys to Arg substitution at the site homologous to felid residue 125 causes constitutive activation of the receptor [31], and the exact same change is thought to be responsible for melanism in the Alaska silver fox [32]; therefore, we consider C125R to be the likely causative alteration in the Geoffroy’s cat.
Transmission of melanism has not been described in these species, but the amino acid substitutions and patterns of association predict that melanism in the pampas cat and kodkod is recessively inherited, and that melanism in the Geoffroy’s cat is dominantly inherited. Frequencies of the melanism alleles are 0.71, 0.5, and 0.15 for the pampas cat, the kodkod, and Geoffroy’s cat, respectively, and are consistent with the field-based assessments of melanism frequency in the populations from which each sample was drawn (Table 1). Like other wild felids that occur at low densities, small sample sizes constrain the power of studies based on estimates of genotype frequency. With that caveat, a relatively high prevalence of melanism due to independent mutations in these three species suggests that the phenotype is not deleterious [33].
CATCH-Seq libraries: construction, analysis, and validation
Originally developed as a cost-effective way for targeted resequencing of human regions that are highly polymorphic, clone-based target capture is also uniquely suited for studying non-model organisms for which large insert genomic libraries from a closely related organism are available. To survey genetic variation within and around ASIP and MC1R in Leopardus spp., we identified a series of fosmids from the domestic cat reference genome that contain and surround each locus (Fig. 2). For ASIP, we used 8 fosmids that contain 299 kb of DNA within an 842 kb interval; for MC1R, we used 5 fosmids that contain 251 kb of DNA within a 409 kb interval (Fig. 2, Table S7 in S1 Data). Illumina paired-end libraries were constructed from sheared genomic DNA of 57 animals (Table S1 in S1 Data). From 54 libraries that passed quality control, material selected by CATCH-Seq using biotinylated probes prepared from the 13 fosmids was sequenced in a multiplexed format; a total of 1,053,331,306 paired-end reads (50 bp) were collected.
Summary alignment and mapping statistics are presented in Table 2. The current domestic cat assembly (Felis_catus-6.2) spans 2.43 Gb and represents ~14x Q20 coverage, but has not yet been fully annotated. Approximately 70% of our sequence reads aligned to the cat genome; of these, ~58% aligned to one of the ASIP or MC1R target fosmids considering the entire sequence of the fosmids, and ~7% aligned to the ASIP or MC1R target considering the repeat-masked sequence of the fosmids, consistent with an enrichment value of ~500–~2500-fold (Table 2, Table S9 in S1 Data).
Fig. 2. Positions of fosmids and neighboring genes at melanism loci. 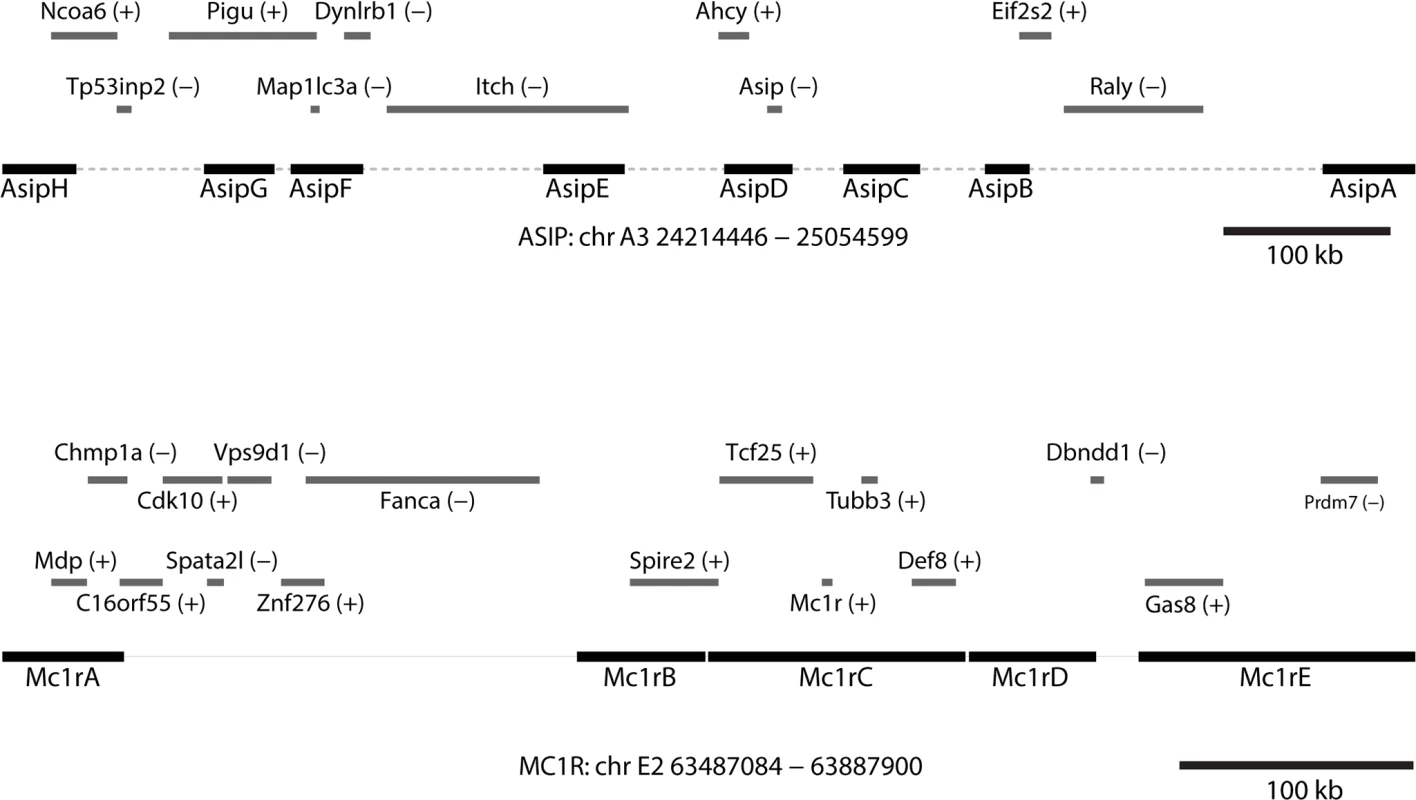
Contig lengths and spacing are drawn to scale (see Table S7 in S1 Data for individual coordinates); sense or antisense transcription, relative to the chromosomal orientation, is indicated with (+)/(-). Tab. 2. Alignment coverage summary for CATCH-Seq libraries. 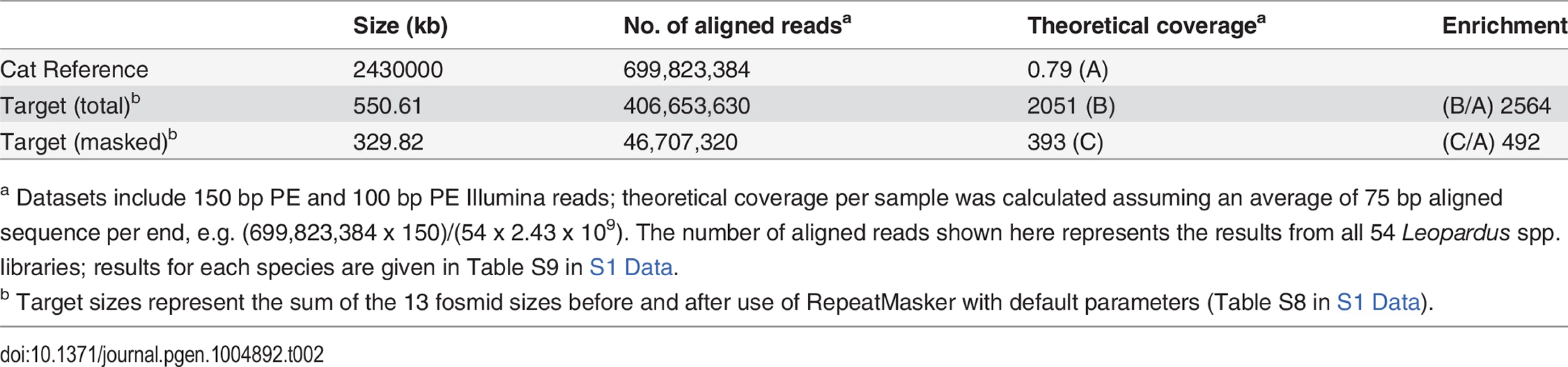
a Datasets include 150 bp PE and 100 bp PE Illumina reads; theoretical coverage per sample was calculated assuming an average of 75 bp aligned sequence per end, e.g. (699,823,384 x 150)/(54 x 2.43 x 109). The number of aligned reads shown here represents the results from all 54 Leopardus spp. libraries; results for each species are given in Table S9 in S1 Data. The mean coverage of each fosmid was 150x, with 71%–99% (mean, 92%) of non-repetitive sequence covered at > 10x (Table S10 in S1 Data). Nucleotide divergence between the domestic cat and Leopardus spp. was ~1–2%, making the domestic cat unsuitable as a reference for variant-calling; therefore, we used SAMtools to develop a consensus sequence for each species separately (combining melanistic and non-melanistic individuals within each species). The species-specific consensus sequence was then used to establish high-confidence variant calls for each individual according to best practices for the Genome Analysis Toolkit, and likely haplotype structure was inferred using Beagle.
We identified 149, 383, and 474 SNPs in the kodkod, pampas cat, and Geoffroy’s cat, respectively, which yield estimates of percent nucleotide diversities of 0.0180 (kodkod), 0.0451 (pampas cat), and 0.0572 (Geoffroy’s cat) (Table S11 in S1 Data). The different nucleotide diversities are not artifacts of ascertainment, since alignment statistics and coverage are similar for the three species (Supplemental Tables S9, S10). (Coverage of fosmids in the Geoffroy’s cat was not as complete, ~15x, as in the kodkod (~35x) or pampas cat (~20x) [Table S10 in S1 Data], but more SNPs were identified in Geoffroy’s cat than in the other two species). Instead, the differences in nucleotide diversity likely reflect species-specific demographic histories as described below.
To help validate our pipeline, and to further explore the pattern of variant distribution between and within species, we carried out a phylogenetic analysis for the ASIP and MC1R regions using a Bayesian Markov Chain Monte Carlo (MCMC) approach [34], in which individual chromosomes for each locus were clustered and the clade credibility for the major nodes was examined. Although this approach does not account for recombination and therefore is not informative with regard to gene genealogy within species, the tree topologies should approximate evolutionary relationships among species, and also provide some assessment of quality control with regard to potential population structure as well as the accuracy of species identification. As depicted in Fig. 3, the posterior clade probability for the nodes representing the divergence of the three species is very high for both ASIP and MC1R loci, with basal relationships that recapitulate the known topology in which the kodkod and Geoffroy’s cat are more closely related to each other than to the pampas cat. For the pampas cat and Geoffroy’s cat, chromosomes bearing melanism mutations (ASIP for pampas cat, MC1R for Geoffroy’s cat) do not cluster together; for the kodkod, there is some clustering of melanistic ASIP chromosomes that may reflect recent population history, as described below.
Fig. 3. Phylogenetic relationships among chromosomes for each locus inferred with BEAST2 [34]. ![Phylogenetic relationships among chromosomes for each locus inferred with BEAST2 [<em class="ref">34</em>].](https://www.prelekara.sk/media/cache/resolve/media_object_image_small/media/image/1ea89f7b152ae5d52f266a499937f579.png)
Posterior probabilistic support for the main branches of the maximal clade credibility trees are: a – 0.987, b – 0.991, c – 0.988, d – 0.988, e – 0.999, f – 0.999, g – 0.995, h – 0.992. Colored squares on the right of each tree indicate whether each haplotype is linked to (e.g. contains) a melanism-associated mutation. Molecular evolution of melanism mutations
A fine-scale view of haplotype patterns and nucleotide diversity allows additional insight into the evolutionary history underlying felid melanism. Positive selection is expected to give rise to reduced nucleotide diversity at the melanism locus (ASIP in the pampas cat and kodkod, MC1R in Geoffroy’s cat), but not at the “control” locus (MC1R in the pampas cat and kodkod, ASIP in Geoffroy’s cat).
Fig. 4 depicts the pattern of variation for individual chromosomes at the ASIP (pampas cat and kodkod) and MC1R (Geoffroy’s cat) loci, grouped according to whether the chromosome carries an ancestral or derivative allele, and whether it was observed in a melanistic (m/m or +/M) or a non-melanistic (+/+ or +/m) animal. At the level of individual haplotypes, multiple variants in the pampas cat define a large haplotype block (Fig. 4A) that extends throughout the AsipD fosmid and that contains the ASIP melanism mutation. Haplotype diversity is also reduced for melanism-bearing chromosomes in the pampas cat: within the AsipD fosmid, there are 3 haplotypes among 26 melanism-bearing chromosomes compared to 8 haplotypes among 10 ancestral chromosomes. In the Geoffroy’s cat, four variants within the Mc1rC fosmid delineate a ~1kb haplotype block almost perfectly associated with melanism (Fig. 4B); beyond this central core, there is no obvious distinction in haplotype structure or haplotype diversity between chromosomes that carry melanism mutations and those that do not. Haplotype patterns in the kodkod reveal striking differences between melanism-bearing and ancestral chromosomes, but relatively little within-group difference; in the AsipD fosmid, there are 3 haplotypes among 10 melanism-bearing chromosomes, and 5 haplotypes among 14 ancestral chromosomes (Fig. 4C).
Fig. 4. Haplotype structures for melanism loci for pampas cat (A), Geoffroy’s cat (B), and kodkod (C), arranged according to whether they carry an ancestral (yellow) or derivative (black) allele at the causative position, and whether they were observed in a non-melanistic (+/+, +/m) or melanistic (m/m, +/M) individual. 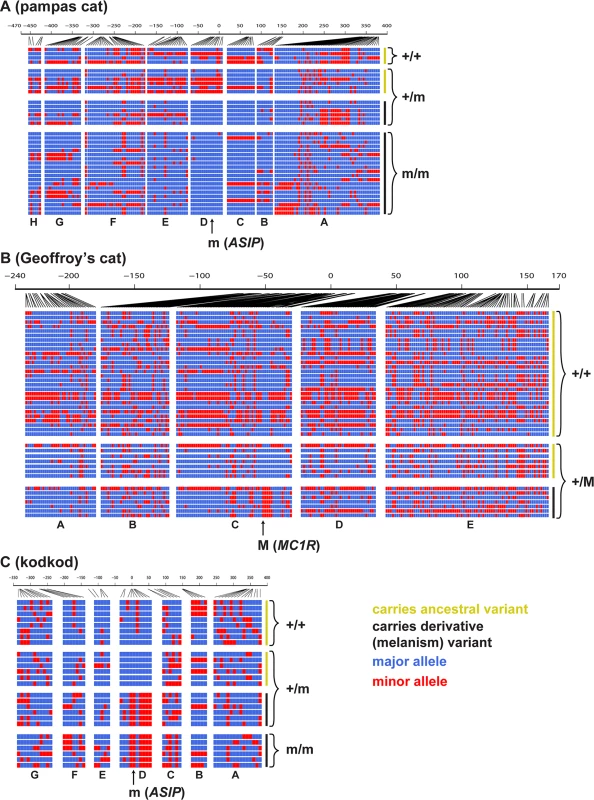
Major and minor alleles are depicted in blue and red respectively, and genomic coordinates are represented in kb upstream or downstream from the causative position. Letters indicating the fosmids that correspond to each targeted segments are shown at the bottom of each plot. We next compared nucleotide diversity between melanism-bearing and ancestral chromosomes (Fig. 5A, B, C), using fosmid origin (Fig. 2) as a window for proximity to the causative variant. In the pampas cat, melanism-bearing chromosomes exhibit a clear signature of selection (Fig. 5A), with reduction of nucleotide diversity that extends across a ~500 kb interval surrounding the causative variant (ASIP fosmids AsipB, AsipC, AsipD, AsipE, AsipF, AsipG), but levels of nucleotide diversity similar to ancestral chromosomes in flanking fosmids (ASIP fosmids AsipA, AsipH). In the Geoffroy’s cat (Fig. 5B), melanism-bearing chromosomes also exhibit a window of reduced nucleotide diversity in a region ~50 kb upstream of the causative variant (MC1R fosmid Mc1rB), although the extent of the difference is not as striking as in the pampas cat. In the kodkod, nucleotide diversity is similarly low for both melanism-bearing and ancestral chromosomes in the AsipD fosmid; in two adjacent fosmids (AsipE, AsipF), levels are modestly higher for melanism-bearing compared to ancestral chromosomes (Fig. 5C).
Fig. 5. Nucleotide diversity calculated for ancestral (yellow) vs. melanism-bearing (black) chromosomes, and for individuals according to their phenotype (melanistic, black vs. non-melanistic, yellow), for pampas cat (A, D), Geoffory’s cat (B, E) and kodkod (C, F). 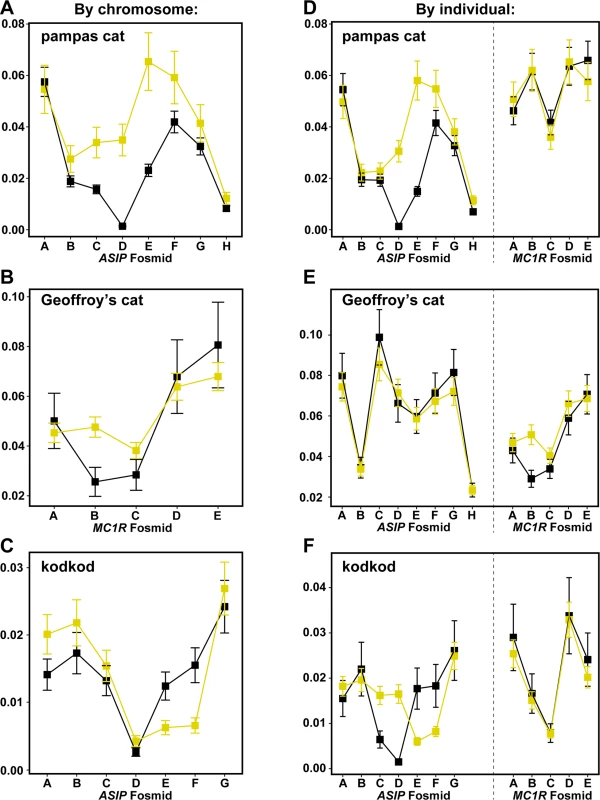
As described in the text, the latter approach allows comparison between loci within each species, illustrating, e.g. that patterns of nucleotide diversity are similar between melanistic individuals for the “control” locus (ASIP in the Geoffroy’s cat, MC1R in the pampas cat and kodkod). Error bars indicate standard error of the mean (SEM). We also calculated nucleotide diversity for melanistic compared to non-melanistic individuals (Fig. 5D, E, F); although less sensitive for evaluating differences in derivative vs. ancestral chromosomes (since non-melanistic pampas cats and kodkods are a mixture of +/m and +/+), this approach allows a direct comparison of nucleotide diversity at a locus unlinked to the causative variant (MC1R for the pampas cat and kodod; ASIP for the Geoffroy’s cat). There are no systematic differences between melanistic and non-melanistic individuals for loci that are unlinked to the causative variant, whereas differences observed for the pampas cat and Geoffroy’s cat in a by-chromosome analysis of the causative locus (Fig. 5A, B) persist in a by-individual analysis of the causative locus (Fig. 5D, E).
A signature of selection in the pampas cat is also readily apparent from graphs of extended haplotype homozygosity (EHH) and haplotype bifurcation diagrams (Fig. 6A); EHH for melanism-bearing chromosomes in the pampas cat extends for hundreds of kilobases with relatively few bifurcations. By contrast, the Geoffroy’s cat and the kodkod exhibit similar EHH for ancestral and derivative chromosomes (Fig. 6B, C).
Fig. 6. Long-range haplotype analysis for pampas cat (A), Geoffory’s cat (B) and kodkod (C) respectively. 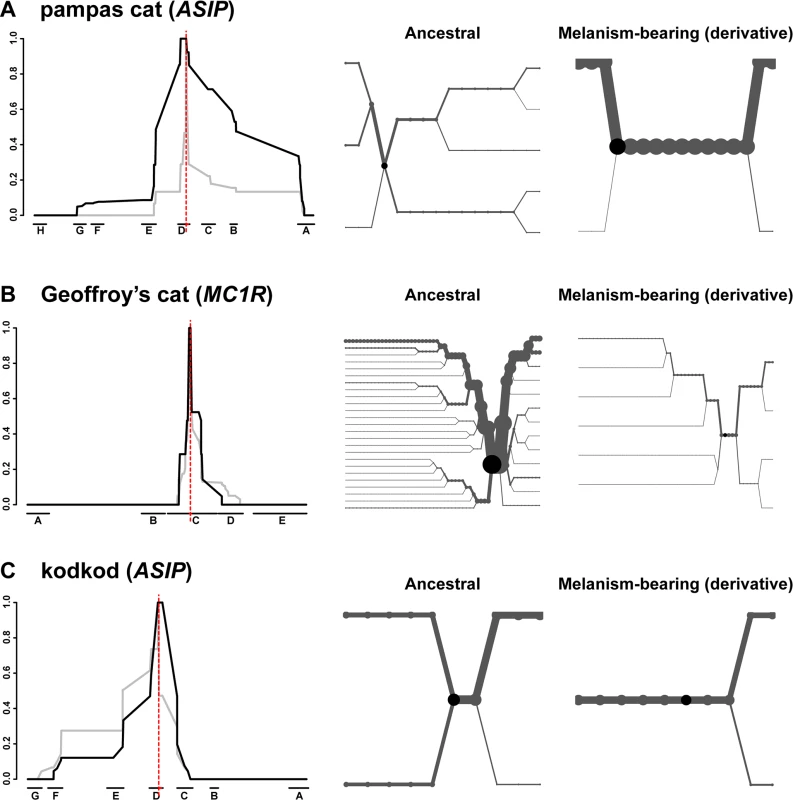
The left-hand side depicts relative EHH according to distance from the causative variant (vertical red line) plotted on the y-axis for ancestral (grey) vs. melanism-bearing (black) chromosomes, as a function of genomic location plotted on the x axis, indicated according to fosmid identity (Fig. 2). The right-hand side depicts haplotype bifurcation plots with origins at the causative variant: branches represent haplotype divergence, and the thickness of the lines is proportional to the number of chromosomes. Haplotype diversity, nucleotide diversity, and EHH in the kodkod are unusual: nucleotide diversity graphs for melanism-bearing and ancestral chromosomes intersect and cross within the AsipD fosmid (Fig. 5C), and the EHH graphs in the kodkod are similar for ancestral and melanism-bearing chromosomes (Fig. 5C). Although this pattern does not lend itself to a simple interpretation, genetic drift is likely to be a contributing factor given the overall reduction of nucleotide diversity; 0.018% in the kodkod compared to 0.045% and 0.057% in the pampas and Geoffroy’s cats, respectively.
Discussion
The application of short-read DNA sequencing technology to longstanding questions in mammalian evolution offers significant opportunities as well as challenges. Morphological variation within a species, or between closely related species, is often hypothesized to serve an adaptive role, a question that can be informed by knowing the molecular basis and evolutionary history of the underlying events. This effort can be challenging, however, in settings where a reference genome does not yet exist. As shown here, our application of a clone-based targeted capture-resequencing strategy reveals distinct evolutionary histories for melanism in three closely related Leopardus species. The results have implications for understanding the biological and genetic basis of melanism in other mammals, inform ongoing efforts in conservation biology, and provide a molecular example that can be applied more widely in population and evolutionary genomics.
Melanism is one of the most common color morphs in domestic and natural populations of birds and mammals, and in most species it is caused by a loss-of-function mutation in ASIP or a gain-of-function mutation in MC1R [3,10,26]. The same now appears to be true in felids; including the three Leopardus species presented here, ASIP or MC1R mutations have now been found to cause melanism in 8 felid species [8,35], a trend that seems likely to continue for the remaining 5 felid species (the jungle cat, the marbled cat, the bobcat, the tigrina, and the serval), in which melanism is recognized but not characterized from a molecular genetic perspective. Loss-of-function mutations in two additional and related pigment type-switching components, Attractin and Mahogunin, cause melanism in the laboratory mouse or rat, but these mutations also cause brain abnormalities and would probably be subject to negative selection in natural populations [36]. In North American wolves, an unusual gain-of-function alteration in beta-defensin 103 causes dominantly inherited melanism and was acquired by hybridization with domestic dogs; however, beta-defensin variants that affect pigmentation have not been recognized outside of dogs, wolves, or coyotes [12,27].
Interspecies hybridization is not uncommon among Leopardus species [19], but we observed no examples of variants that were shared between species. Our results show that at least three independent melanism mutations have occurred during recent evolution of this genus, and have remained polymorphic within each species. Molecular genetic evidence for an adaptive role is strongest for the pampas cat, in which the causative ASIP variant lies on a single major haplotype that extends for hundreds of kilobases as part of a selective sweep. However, absence of long haplotype patterns that clearly distinguish ancestral and melanism-bearing chromosomes in the Geoffroy’s cat and kodkod does not exclude the possibility that melanism is adaptive in these species. The Geoffroy’s cat in particular exhibits patterns of variation that deviate from neutral expectation, with all melanism-bearing chromosomes carrying three non-synonymous mutations in MC1R-coding sequence, and reduced levels of nucleotide diversity in a region ~50 kb upstream of MC1R. Potential selection for melanism in the Geoffroy’s cat may have occurred earlier than in the pampas cat; alternatively, or in addition, a potentially higher recombination rate at MC1R than at ASIP may have diminished the size of an extended haplotype in the Geoffroy’s cat. Studies of additional Geoffroy’s cat populations may help reveal whether the unusual haplotype pattern associated with melanism reflects a specialized population history in Southern Brazil or a more general aspect of the derivative chromosome.
In the kodkod, haplotype structure is similar between melanism-bearing and ancestral chromosomes, but the patterns of nucleotide diversity are unusual. Furthermore, within the fosmid that carries the causative variant (AsipD), multiple SNPs sort the melanism-bearing and ancestral chromosomes into two different lineages. A recent population bottleneck associated with colonization of Chiloé Island could account for the latter observation, but does not easily explain why ancestral chromosomes exhibit a local reduction of nucleotide diversity anchored by the ASIP gene. One intriguing possibility is frequency-dependent balancing selection, such that both melanistic and non-melanistic phenotypes have adaptive value, but only at low frequencies. Larger sample sizes and evaluation of multiple loci would provide additional insight on this hypothesis.
The kodkod is also striking for its very low nucleotide diversity (0.018%), among the lowest described among all mammals [37], which is consistent with assessments from ecological studies indicating that the restricted geographic range of this species coupled with deforestation of Central-southern Chile has reduced its effective population size [14–17,38]. Although it can be challenging to distinguish if loss of genetic diversity is a cause or consequence in any natural population, it is directly related to population reduction.
Melanism is a quintessential example of natural selection in many animals; mechanisms that underlie adaptation can be difficult to discern in larger mammals, although recent work from Allen et al. [9] provides evidence for disruptive selection. In felids, crypsis, presumably as an aid to predation, is thought to drive variation in color patterns, but variation in base color is often postulated to facilitate temperature regulation or response to UV radiation. The latter two mechanisms are oft-cited explanations for Gloger’s rule (correlation between prevalence of darker coloration and equatorial proximity), but are unlikely to explain persistent polymorphism of melanism within Leopardus spp.
Our results point to an important role for natural selection in the evolution of melanism for at least one of the analyzed species, but reveal very different genomic signals that likely reflect unique demographic histories, influenced by different selective processes in addition to drift and recombination. Application of similar approaches to other species will extend our knowledge of how natural selection has shaped color variation, and offer new avenues to investigate the evolutionary history of phenotypic diversity.
Materials and Methods
Animals and genomes
We studied a total of 57 individuals (18 pampas cat, 16 kodkod, and 23 Geoffroy’s cat) whose origin and phenotype are listed in Table S1 in S1 Data. Biological material was either blood (collected from wild-caught individuals in the context of field ecology studies) or tissue (from road-killed animals encountered during routine wildlife surveys); in all cases, sampling was performed following appropriate national regulations for handling animals and biological materials. DNA was prepared by extraction with phenol/chloroform, and assessed for concentration and quality by fluorometry and agarose gel electrophoresis, respectively.
When this work was initiated, we made use of an annotated but incomplete (1.9x) genome assembly of the domestic cat, felCat3 [39]. During the course of the work, two additional assemblies became available, and all coordinates used in the manuscript now refer to felCat5/Felis_catus-6.2, http://genome.wustl.edu/genomes/detail/felis-catus/.
Candidate gene genotyping
After PCR amplification, protein-coding exons of ASIP [8] and MC1R (Table S2 in S1 Data) were analyzed by automated capillary sequencing. We first examined 8 individuals of each species (4 of each phenotype), then extended those results to all available samples (Supplemental Tables S1, S4, S5, S6). Sequencing electropherograms were verified and corrected with Sequencher 4.2 (GeneCodes Corporation), and every potential variant was carefully inspected for confirmation. Homologous nucleotide and amino acid sequences of each gene from additional mammalian species were obtained from GenBank (Table S3 in S1 Data) and aligned using ClustalW.
Targeted resequencing
DNA libraries for Illumina sequencing were generated using standard protocols. Briefly, 700ng to 2.5ug of input DNA (depending on the level of DNA degradation assessed by agarose gel electrophoresis before shearing) was sheared to a size range of 100–500 bp, and custom inline barcodes were added during adapter ligation for paired-end sequencing.
For target selection, we utilized a set of fosmid libraries that had been previously end-sequenced and mapped to a 3x draft assembly, V17e/felCat4 [40]; identity of all fosmids was verified by PCR of each end.
A detailed protocol for probe preparation, hybridization, and capture is described elsewhere (Day et al., manuscript submitted). Briefly, fosmids were pooled according to their inferred individual mass, a total of 1.5ug of input was sheared to 100–500 bp, and DNA fragments were used to prepare biotinylated RNA with a Megascript T7 kit (Ambion). After DNAse treatment and removal of unincorporated nucleotides, hybridizations were carried out in a volume of 26ul that contained 300 ng of probe and 500ng of library DNA (125ng each of 4 inline barcoded libraries). Selected DNA was recovered with magnetic beads and amplified by PCR (20 cycles using standard Illumina primers). Library 4-plex pools were sequenced as 12-plex library sets on a single Illumina HiSeq 2000 lane.
Bioinformatics
We used ASIP and MC1R regions from chromosomes A3 and E2, respectively, as reference sequences, extracted from the Felis_catus-6.2 assembly. The genomic reference was masked for transposable elements and low complexity regions with RepeatMasker [41]. To minimize genotyping errors related to inter-species differences in allele structure and distribution, we first mapped all raw sequence reads against the domestic cat reference using the Burrows-Wheeler Aligner (BWA) with default parameters [42], then used SAMtools [43] to compute consensus sequences for these preliminary alignments, and to de novo assemble targeted regions in each of the three species separately.
Sequence reads were then remapped to these de novo species-specific consensus sequences and subsequently used to for variant calling according to GATK best practices guidelines [44]. Briefly, we started by applying alignment quality control procedures available in GATK to detect sequence intervals with low quality mappings (i.e. possibly related to the presence of sequence variants, such as small insertions or deletions, in subsets of the analyzed samples). In all such cases, a thorough local realignment of the reads was performed to minimize the number of mismatching bases. The GATK Unified Genotyper (UG) tool was applied on realigned reads to infer the genotype structure simultaneously across all samples for each species. The raw genotype calls were subjected to a filtering procedure by imposing thresholds on a set of quality criteria, including minor allele frequency MAF>10%, Phred-scaled mapping quality MQ>40, UG quality by depth QD>2 and UG HaplotypeScore>13. The filtered calls were further restricted to a small subset of high quality calls, to satisfy an average variant density threshold of ~1 per 1kb of target sequence. Finally, the BEAGLE genetic analysis software package [45] was used to check genotype consistency across all samples of each species and infer the haplotype phase of selected variants.
Curated genotype and haplotype data was then used for downstream analyses, including nucleotide diversity and polymorphism statistics with DnaSP 5.10 [46], EHH [47], and generation of median-joining haplotype networks (Network 4.5.0.0; http://www.fluxus-engineering.com). Haplotype bifurcation plots in Fig. 5 were generated with Sweep 1.1 (http://www.broadinstitute.org/mpg/sweep/index.html) and rehh 1.0 [48].
Data access
DNA sequences reported in this manuscript are publicly available from DRYAD (doi:10.5061/dryad.pq482).
Supporting Information
Zdroje
1. Caro TIM (2005) The Adaptive Significance of Coloration in Mammals. BioScience 55 : 125–136.
2. Hoekstra HE (2006) Genetics, development and evolution of adaptive pigmentation in vertebrates. Heredity (Edinb) 97 : 222–234. 16823403
3. Hubbard JK, Uy JA, Hauber ME, Hoekstra HE, Safran RJ (2010) Vertebrate pigmentation: from underlying genes to adaptive function. Trends Genet 26 : 231–239. doi: 10.1016/j.tig.2010.02.002 20381892
4. Johnson WE, Eizirik E, Pecon-Slattery J, Murphy WJ, Antunes A et al. (2006) The late Miocene radiation of modern Felidae: a genetic assessment. Science 311 : 73–77. 16400146
5. Werdelin L, Yamaguchi N, Johnson WE, O’Brien SJ (2010) Phylogeny and evolution of cats (Felidae). In: Macdonald DW, Loveridge AJ, editors. Biology and conservation of wild felids. Oxford; New York: Oxford University Press. pp. 59–82.
6. Robinson R (1990) Homologous genetic variation in the Felidae. Genetica 46 : 1–31.
7. Sunquist ME, Sunquist F (2002) Wild Cats of the World. Chicago, IL: The University of Chicago Press. 25057650
8. Schneider A, David VA, Johnson WE, O’Brien SJ, Barsh GS et al. (2012) How the Leopard Hides Its Spots: ASIP Mutations and Melanism in Wild Cats. PLoS One 7: e50386. doi: 10.1371/journal.pone.0050386 23251368
9. Allen WL, Cuthill IC, Scott-Samuel NE, Baddeley R (2011) Why the leopard got its spots: relating pattern development to ecology in felids. Proc Biol Sci 278 : 1373–1380. doi: 10.1098/rspb.2010.1734 20961899
10. Majerus ME, Mundy NI (2003) Mammalian melanism: natural selection in black and white. Trends Genet 19 : 585–588. 14585605
11. Majerus MEN, Brakefield PM (1998) Melanism: evolution in action. Oxford, NY: Oxford University Press. 25506963
12. Anderson TM, vonHoldt BM, Candille SI, Musiani M, Greco C et al. (2009) Molecular and evolutionary history of melanism in North American gray wolves. Science 323 : 1339–1343. doi: 10.1126/science.1165448 19197024
13. Kawanishi K, Sunquist ME, Eizirik E, Lynam AJ, Ngoprasert D et al. (2010) Near fixation of melanism in leopards of the Malay Peninsula. Journal of Zoology 282 : 201–206.
14. Dunstone N, Durbin L, Wyllie I, Freer R, Jamett GA et al. (2002) Spatial organization, ranging behaviour and habitat use of the kodkod (Oncifelis guigna) in southern Chile. Journal of Zoology 257 : 1–11.
15. Napolitano CG (2012) Filogeografía, inferencia demográfica y genética de la conservación del felino Leopardus guigna en el sur de sudamérica. Programa de doctorado en Ciencias Silvoagropecuarias y Veterinarias, Universidad de Chile 255 pp.
16. Napolitano C, Johnson W, Sanderson J, O,ÄôBrien S, Rus H, A et al. (2014) Phylogeography and population history of Leopardus guigna, the smallest American felid. Conserv Genet 15 : 631–653.
17. Sanderson J, Sunquist M.E., Iriarte A.W. (2002) Natural history and landscape-use of guignas (Oncifelis guigna) on Isla Grande de Chiloe, Chile. Journal of Mammalogy 608–613.
18. Trigo TC, Freitas TR, Kunzler G, Cardoso L, Silva JC et al. (2008) Inter-species hybridization among Neotropical cats of the genus Leopardus, and evidence for an introgressive hybrid zone between L. geoffroyi and L. tigrinus in southern Brazil. Mol Ecol 17 : 4317–4333. doi: 10.1111/j.1365-294X.2008.03919.x 18785898
19. Trigo TC, Schneider A, de Oliveira TG, Lehugeur LM, Silveira L et al. (2013) Molecular data reveal complex hybridization and a cryptic species of neotropical wild cat. Curr Biol 23 : 2528–2533. doi: 10.1016/j.cub.2013.10.046 24291091
20. George RD, McVicker G, Diederich R, Ng SB, MacKenzie AP et al. (2011) Trans genomic capture and sequencing of primate exomes reveals new targets of positive selection. Genome Research 21 : 1686–1694. doi: 10.1101/gr.121327.111 21795384
21. Day K, Song J, Absher D (2014) Targeted sequencing of large genomic regions with CATCH-Seq. PloS One Unpublished: (in review).
22. Klungland H, Vage DI (2003) Pigmentary switches in domestic animal species. Ann N Y Acad Sci 994 : 331–338. 12851333
23. Kaelin CB, Barsh GS (2012) Molecular Genetics of Coat Colour, Texture and Length in the Dog. In: Ostrander EA, Ruvinsky A, editors. The Genetics of the Dog, 2nd ed. Boston, MA: CABI. pp. 57–80.
24. Simon JD, Peles D, Wakamatsu K, Ito S (2009) Current challenges in understanding melanogenesis: bridging chemistry, biological control, morphology, and function. Pigment Cell Melanoma Res 22 : 563–579. doi: 10.1111/j.1755-148X.2009.00610.x 19627559
25. Jackson IJ (1994) Molecular and developmental genetics of mouse coat color. Annu Rev Genet 28 : 189–217. 7893123
26. Suzuki H (2013) Evolutionary and phylogeographic views on Mc1r and Asip variation in mammals. Genes Genet Syst 88 : 155–164. 24025244
27. Candille SI, Kaelin CB, Cattanach BM, Yu B, Thompson DA et al. (2007) A beta-defensin mutation causes black coat color in domestic dogs. Science 318 : 1418–1423. 17947548
28. Gunn TM, Miller KA, He L, Hyman RW, Davis RW et al. (1999) The mouse mahogany locus encodes a transmembrane form of human attractin. Nature 398 : 152–156. 10086356
29. He L, Lu XY, Jolly AF, Eldridge AG, Watson SJ et al. (2003) Spongiform degeneration in mahoganoid mutant mice. Science 299 : 710–712. 12560552
30. McNulty JC, Jackson PJ, Thompson DA, Chai B, Gantz I et al. (2005) Structures of the agouti signaling protein. J Mol Biol 346 : 1059–1070. 15701517
31. Robbins LS, Nadeau JH, Johnson KR, Kelly MA, Roselli-Rehfuss L et al. (1993) Pigmentation phenotypes of variant extension locus alleles result from point mutations that alter MSH receptor function. Cell 72 : 827–834. 8458079
32. Vage DI, Lu D, Klungland H, Lien S, Adalsteinsson S et al. (1997) A non-epistatic interaction of agouti and extension in the fox, Vulpes vulpes. Nat Genet 15 : 311–315. 9054949
33. Gray SM, McKinnon JS (2007) Linking color polymorphism maintenance and speciation. Trends Ecol Evol 22 : 71–79. 17055107
34. Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29 : 1969–1973. doi: 10.1093/molbev/mss075 22367748
35. Eizirik E, Yuhki N, Johnson WE, Menotti-Raymond M, Hannah SS et al. (2003) Molecular genetics and evolution of melanism in the cat family. Curr Biol 13 : 448–453. 12620197
36. He L, Eldridge AG, Jackson PK, Gunn TM, Barsh GS (2003) Accessory proteins for melanocortin signaling: attractin and mahogunin. Ann N Y Acad Sci 994 : 288–298. 12851328
37. Leffler EM, Bullaughey K, Matute DR, Meyer WK, Segurel L et al. (2012) Revisiting an old riddle: what determines genetic diversity levels within species? PLoS Biol 10: e1001388. doi: 10.1371/journal.pbio.1001388 22984349
38. Echeverria C, Coomes D, Salas J, Rey-Benayas JM, Lara A et al. (2006) Rapid deforestation and fragmentation of Chilean Temperate Forests. Biological Conservation 130 : 481–494.
39. Pontius JU, O’Brien SJ (2007) Genome Annotation Resource Fields—GARFIELD: a genome browser for Felis catus. J Hered 98 : 386–389. 17646276
40. Mullikin JC, Hansen NF, Shen L, Ebling H, Donahue WF et al. (2010) Light whole genome sequence for SNP discovery across domestic cat breeds. BMC Genomics 11 : 406. doi: 10.1186/1471-2164-11-406 20576142
41. Zhi D, Raphael BJ, Price AL, Tang H, Pevzner PA (2006) Identifying repeat domains in large genomes. Genome Biol 7: R7. 16507140
42. Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 : 1754–1760. doi: 10.1093/bioinformatics/btp324 19451168
43. Li H, Handsaker B, Wysoker A, Fennell T, Ruan J et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25 : 2078–2079. doi: 10.1093/bioinformatics/btp352 19505943
44. Genome Sequencing and Analysis Group (2012) Best Practice Variant Detection with the GATK v3. Cambridge, MA: Broad Institute. doi: 10.1111/cob.12003 23271062
45. Browning SR, Browning BL (2007) Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 81 : 1084–1097. 17924348
46. Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25 : 1451–1452. doi: 10.1093/bioinformatics/btp187 19346325
47. Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ et al. (2002) Detecting recent positive selection in the human genome from haplotype structure. Nature 419 : 832–837. 12397357
48. Gautier M, Vitalis R (2012) rehh: an R package to detect footprints of selection in genome-wide SNP data from haplotype structure. Bioinformatics 28 : 1176–1177. doi: 10.1093/bioinformatics/bts115 22402612
Štítky
Genetika Reprodukčná medicína
Článek 2014 Reviewer Thank YouČlánek Closing the Gap between Knowledge and Clinical Application: Challenges for Genomic TranslationČlánek Discovery of CTCF-Sensitive Cis-Spliced Fusion RNAs between Adjacent Genes in Human Prostate CellsČlánek K-homology Nuclear Ribonucleoproteins Regulate Floral Organ Identity and Determinacy in ArabidopsisČlánek A Nitric Oxide Regulated Small RNA Controls Expression of Genes Involved in Redox Homeostasis inČlánek Contribution of the Two Genes Encoding Histone Variant H3.3 to Viability and Fertility in MiceČlánek The Genetic Architecture of the Genome-Wide Transcriptional Response to ER Stress in the Mouse
Článok vyšiel v časopisePLOS Genetics
Najčítanejšie tento týždeň
2015 Číslo 2- Gynekologové a odborníci na reprodukční medicínu se sejdou na prvním virtuálním summitu
- Je „freeze-all“ pro všechny? Odborníci na fertilitu diskutovali na virtuálním summitu
-
Všetky články tohto čísla
- 2014 Reviewer Thank You
- Systematic Cell-Based Phenotyping of Missense Alleles Empowers Rare Variant Association Studies: A Case for and Myocardial Infarction
- African Glucose-6-Phosphate Dehydrogenase Alleles Associated with Protection from Severe Malaria in Heterozygous Females in Tanzania
- Genomics of Divergence along a Continuum of Parapatric Population Differentiation
- microRNAs Regulate Cell-to-Cell Variability of Endogenous Target Gene Expression in Developing Mouse Thymocytes
- A Rolling Circle Replication Mechanism Produces Multimeric Lariats of Mitochondrial DNA in
- Closing the Gap between Knowledge and Clinical Application: Challenges for Genomic Translation
- Partially Redundant Enhancers Cooperatively Maintain Mammalian Expression Above a Critical Functional Threshold
- Discovery of Transcription Factors and Regulatory Regions Driving Tumor Development by ATAC-seq and FAIRE-seq Open Chromatin Profiling
- Mutations in Result in Ocular Coloboma, Microcornea and Cataracts
- A Genome-Wide Hybrid Incompatibility Landscape between and
- Recurrent Evolution of Melanism in South American Felids
- Discovery of CTCF-Sensitive Cis-Spliced Fusion RNAs between Adjacent Genes in Human Prostate Cells
- Tissue Expression Pattern of PMK-2 p38 MAPK Is Established by the miR-58 Family in
- Essential Role for Endogenous siRNAs during Meiosis in Mouse Oocytes
- Matrix Metalloproteinase 2 Is Required for Ovulation and Corpus Luteum Formation in
- Evolutionary Signatures amongst Disease Genes Permit Novel Methods for Gene Prioritization and Construction of Informative Gene-Based Networks
- RR-1 Cuticular Protein TcCPR4 Is Required for Formation of Pore Canals in Rigid Cuticle
- GC-Content Evolution in Bacterial Genomes: The Biased Gene Conversion Hypothesis Expands
- Proteotoxic Stress Induces Phosphorylation of p62/SQSTM1 by ULK1 to Regulate Selective Autophagic Clearance of Protein Aggregates
- K-homology Nuclear Ribonucleoproteins Regulate Floral Organ Identity and Determinacy in Arabidopsis
- A Nitric Oxide Regulated Small RNA Controls Expression of Genes Involved in Redox Homeostasis in
- HYPER RECOMBINATION1 of the THO/TREX Complex Plays a Role in Controlling Transcription of the Gene in Arabidopsis
- Mitochondrial and Cytoplasmic ROS Have Opposing Effects on Lifespan
- Structured Observations Reveal Slow HIV-1 CTL Escape
- An Integrative Multi-scale Analysis of the Dynamic DNA Methylation Landscape in Aging
- Combining Natural Sequence Variation with High Throughput Mutational Data to Reveal Protein Interaction Sites
- Transhydrogenase Promotes the Robustness and Evolvability of Deficient in NADPH Production
- Regulators of Autophagosome Formation in Muscles
- Genomic Selection and Association Mapping in Rice (): Effect of Trait Genetic Architecture, Training Population Composition, Marker Number and Statistical Model on Accuracy of Rice Genomic Selection in Elite, Tropical Rice Breeding Lines
- Eye Selector Logic for a Coordinated Cell Cycle Exit
- Inflammation-Induced Cell Proliferation Potentiates DNA Damage-Induced Mutations
- The DNA Polymerase δ Has a Role in the Deposition of Transcriptionally Active Epigenetic Marks, Development and Flowering
- Contribution of the Two Genes Encoding Histone Variant H3.3 to Viability and Fertility in Mice
- Membrane Recognition and Dynamics of the RNA Degradosome
- P-TEFb, the Super Elongation Complex and Mediator Regulate a Subset of Non-paused Genes during Early Embryo Development
- is a Long Non-coding RNA in JNK Signaling in Epithelial Shape Changes during Drosophila Dorsal Closure
- A Pleiotropy-Informed Bayesian False Discovery Rate Adapted to a Shared Control Design Finds New Disease Associations From GWAS Summary Statistics
- Genome-wide Association Study Identifies Shared Risk Loci Common to Two Malignancies in Golden Retrievers
- and Hyperdrive Mechanisms (in Mouse Meiosis)
- Elevated In Vivo Levels of a Single Transcription Factor Directly Convert Satellite Glia into Oligodendrocyte-like Cells
- Systemic Delivery of MicroRNA-101 Potently Inhibits Hepatocellular Carcinoma by Repressing Multiple Targets
- Pooled Sequencing of 531 Genes in Inflammatory Bowel Disease Identifies an Associated Rare Variant in and Implicates Other Immune Related Genes
- Abscission Is Regulated by the ESCRT-III Protein Shrub in Germline Stem Cells
- Temperature Stress Mediates Decanalization and Dominance of Gene Expression in
- Transcriptome Wide Annotation of Eukaryotic RNase III Reactivity and Degradation Signals
- The Exosome Component Rrp6 Is Required for RNA Polymerase II Termination at Specific Targets of the Nrd1-Nab3 Pathway
- Sex-specific -regulatory Variation on the X Chromosome
- Regulation of Toll-like Receptor Signaling by the SF3a mRNA Splicing Complex
- Modeling of the Human Alveolar Rhabdomyosarcoma Chromosome Translocation in Mouse Myoblasts Using CRISPR-Cas9 Nuclease
- Asymmetry of the Budding Yeast Tem1 GTPase at Spindle Poles Is Required for Spindle Positioning But Not for Mitotic Exit
- TIM Binds Importin α1, and Acts as an Adapter to Transport PER to the Nucleus
- Antagonistic Roles for KNOX1 and KNOX2 Genes in Patterning the Land Plant Body Plan Following an Ancient Gene Duplication
- The Genetic Architecture of the Genome-Wide Transcriptional Response to ER Stress in the Mouse
- Fatty Acid Synthase Cooperates with Glyoxalase 1 to Protect against Sugar Toxicity
- Region-Specific Activation of mRNA Translation by Inhibition of Bruno-Mediated Repression
- An Essential Role of the Arginine Vasotocin System in Mate-Guarding Behaviors in Triadic Relationships of Medaka Fish ()
- Interaction between the tRNA-Binding and C-Terminal Domains of Yeast Gcn2 Regulates Kinase Activity In Vivo
- Hyper-Acetylation of Histone H3K56 Limits Break-Induced Replication by Inhibiting Extensive Repair Synthesis
- Prodomain Removal Enables Neto to Stabilize Glutamate Receptors at the Neuromuscular Junction
- Recent Selective Sweeps in North American Show Signatures of Soft Sweeps
- Identification and Functional Analysis of Healing Regulators in
- A Multi-Megabase Copy Number Gain Causes Maternal Transmission Ratio Distortion on Mouse Chromosome 2
- Drosophila Casein Kinase I Alpha Regulates Homolog Pairing and Genome Organization by Modulating Condensin II Subunit Cap-H2 Levels
- The Hippo Pathway Regulates Homeostatic Growth of Stem Cell Niche Precursors in the Ovary
- PLOS Genetics
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Genomic Selection and Association Mapping in Rice (): Effect of Trait Genetic Architecture, Training Population Composition, Marker Number and Statistical Model on Accuracy of Rice Genomic Selection in Elite, Tropical Rice Breeding Lines
- Discovery of Transcription Factors and Regulatory Regions Driving Tumor Development by ATAC-seq and FAIRE-seq Open Chromatin Profiling
- Evolutionary Signatures amongst Disease Genes Permit Novel Methods for Gene Prioritization and Construction of Informative Gene-Based Networks
- Proteotoxic Stress Induces Phosphorylation of p62/SQSTM1 by ULK1 to Regulate Selective Autophagic Clearance of Protein Aggregates
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



