-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The effect of docetaxel on molecular melting profile of DNA extracted from human breast adenocarcinoma MCF-7 cells
Aim:
The aim of this study was to evaluate the genotoxic effect of docetaxel on the human adenocarcinoma MCF-7 cells to detect genetic variations and discover potential associations between the drug and the genotoxic damage of MCF-7 cells. Materials and Methods: High resolution melt analysis (HRM) of genomic DNA isolated from MCF-7 cells was used. Two primers of MDR1 gene were selected: 5´-TGGGGCTTTTAGTGTTGGAC-3´, 5´-TGTGGAGAGCTGGATAAAGTCA-3´.Results:
The significant alterations in the melting temperature Tm of DNA treated with docetaxel in the concentrations of 1 µmol/l and 250 µmol/l were observed. Comparing G+C/A+T ratios the increase of the relative content of G, C was detected. Minor changes of the nucleotide content were observed when compared the sequences of DNA for the control and the docetaxel treated group.Conclusions:
Using MDR1 primer pairs, our results confirmed that MCF-7 cells are susceptible to genomic DNA instability when exposed to docetaxel.Keywords:
MCF-7 cell line, high-resolution melting analysis, docetaxel
Authors: Marianna Trebuňová 1; Ján Rosocha 1; Galina Laputková 2; Mario Jančošek 3; Jozef Živčák 4
Authors place of work: Associated Tissue Bank UNLP Košice, Rastislavova 4 , 0 1 90 Košice, Slovakia 1; Department of Medical and Clinical Biophysics, Faculty of Medicine, P. J. Šafarik University 2; University of Presov in Presov, 17. novembra 15, 080 01 Prešov, Slovakia 3; Faculty of Mechanical Engineering, Department of Biomedical Engineering and Measurement, Technical University, Letná 9, Košice, Slovak Republic 4; Trieda SNP 1, Košice, 00 11, Slovakia 4
Published in the journal: Lékař a technika - Clinician and Technology No. 1, 2014, 44, 43-48
Category: Původní práce
Summary
Aim:
The aim of this study was to evaluate the genotoxic effect of docetaxel on the human adenocarcinoma MCF-7 cells to detect genetic variations and discover potential associations between the drug and the genotoxic damage of MCF-7 cells. Materials and Methods: High resolution melt analysis (HRM) of genomic DNA isolated from MCF-7 cells was used. Two primers of MDR1 gene were selected: 5´-TGGGGCTTTTAGTGTTGGAC-3´, 5´-TGTGGAGAGCTGGATAAAGTCA-3´.Results:
The significant alterations in the melting temperature Tm of DNA treated with docetaxel in the concentrations of 1 µmol/l and 250 µmol/l were observed. Comparing G+C/A+T ratios the increase of the relative content of G, C was detected. Minor changes of the nucleotide content were observed when compared the sequences of DNA for the control and the docetaxel treated group.Conclusions:
Using MDR1 primer pairs, our results confirmed that MCF-7 cells are susceptible to genomic DNA instability when exposed to docetaxel.Keywords:
MCF-7 cell line, high-resolution melting analysis, docetaxelInstroduction
Systematic chemotherapy and hormonal therapy is the most essential factor in reducing mortality in breast cancer patients. Among diverse chemotherapies, docetaxel provides both symptomatic and survival benefits in early breast cancer patients but did not improve survival in metastatic breast cancer trials [1]. The response rate to docetaxel is 40-60 % even in first-line chemotherapy and it decreases to 20-30 % in the second-or third-line chemotherapy [2]. Docetaxel is a semi-synthetic, second-generation taxane derived from a compound found in the yew tree Taxus baccata [3]. Taxanes exert their effects by binding to β-tubulin subunits of microtubules. By stabilizing microtubules, docetaxel prevents the dynamic polymerization/de-polymerization processes essential during mitosis [4]. In this way, apoptosis is induced by blocking cell growth in the G2-M phase by disrupting normal mitotic spindle formation. Moreover, additional signal transduction pathways involved in apoptosis have also been suggested [5, 6]. Caspase-3 actively participates in apoptosis of various tumor cells [7] and activation of caspase-3-dependent pathway has been also regarded as the triggering event of apoptosis induced by taxanes. In addition, it was shown that the treatment with docetaxel leads to DNA fragmentation [8, 9].
Furthermore, mechanisms of sensitivity or resistance to anticancer chemotherapy still remains not fully understood. Numerous tumor drug resistance mechanisms have been demonstrated that attempt to explain mechanisms involved in the resistance to chemotherapeutic agents such as decreased drug transport inside the tumor cells or enhanced drug efflux, increased capability of cancer cells to stop cycling and increased tolerance to drug-induced damage to DNA [10, 11].
Multidrug resistance, in which cancerous tissues exhibit decreased sensitivity to diverse drugs is by far the most studied form of anticancer drug resistance. Overexpression of P-glycoprotein (P-gp) – the product of the multidrug resistance 1 (MDR1/ABCB1) gene, in cancer cells resistant to a variety of anti-tumour alkaloids and antibiotics confirmed clinically is one of the best studied and characterized. However, P-gp itself may have a direct influence on the function of proteins involved in regulatory pathways, including apoptotic progression (such as p53, caspase-3 and pokemon) [12, 13]. A growing number of preclinical and clinical studies have demonstrated that polymorphism of the MDR1 gene may be a factor in the overall outcome of pharmacotherapy for numerous diseases [7].
High resolution melting (HRM) analysis is an attractive platform to cope with broad spectrum of human genetic mutations [14] and polymorphisms [15] as it is single step, closed tube, and cost-effective. HRM includes the precise monitoring of the variation in fluorescence initiated by the release of an intercalating DNA dye from a DNA duplex as it is denatured by increasing temperature. The accuracy of the melt curve is maximized by acquiring fluorescence data over as low temperature increments as 0.01°C.
Considering the fact that the low chemotherapeutic efficiency in cancer patients is related with resistance of cytostatic agents and DNA belongs to the main targets of many conventional anticancer agents, the aim of our study was to evaluate the genotoxic effects of docetaxel to detect potential associations between the drug sensitivities and the genotoxic damage of MCF-7 cell line compared with control untreated MCF-7 cells using HRM analysis. Likewise, real-time polymerase chain reaction analyses were selected to determine the genetic DNA expression levels of the MDR1 gene.
Materials and Metods
Cell Growth Conditions and Cell Passaging
The MCF-7 cell line was purchased from the American Type Culture Collection (ATCC, USA). Cells were grown according to the procedure of Trebuňová et al. 2012 [16]. Briefly, a monolayer of cells were cultured in 10% Dulbecco's Modified Eagle Medium (Invitrogen, USA) containing fetal bovine serum (FBS, Invitrogen), mixture of 5000 units of penicillin and 5000 µg of streptomycin/ml in 0.85 % saline (PenStrep) (Invitrogen, USA). Cells were kept in the Heracell 150i incubator (Thermo Scientific, USA) at 37 °C in 5 % CO2 and 95 % humidity. All solutions used were of the cell culture grade quality. The equipment employed was commercially pre-sterilized and disposable.
Drugs
Docetaxel were kindly provided by Dr. Andrašina (UPJS Kosice, Faculty of Medicine, Department of Oncology). Drug was stored at room temperature in polysorbate 80 as 40 mg/ml concentrate. The solutions of cytostatics were vigorously stirred before dilution in the filtered cell culture medium and stored at 20°C. Cytostatics were directly applied into the culture medium to obtain the final concentrations 1 µmol/l, 3 µmol/l, 50 µmol/l, 100 µmol/l, and 250 µmol/l. The cells were allowed to grow for 2.5 hours and were subsequently treated with fresh medium, after which point they were incubated for 60 hours at 37 °C. After 60 h of exposure, the cells were three times washed with phosphate buffer saline (PBS), pelleted and stored at -80 °C.
DNA samples
DNA samples were extracted from both non-affected and docetaxel-treated MCF-7 cells. Pellets were resuspended in lysis buffer and DNA was isolated according to the manufacturer´s protocol (SensiFASTTM SYBR, No-ROX Kit, Bioline, UK). The concentration of DNA was determined spectrophotometrycally with NanoPhotometerTM (Implen, Germany): MCF-7/c - 78 ng/ml, MCF-7/1 - 63 ng/ml, MCF-7/250 - 62 ng/ml.
Melting analysis
Two primers (Sigma, USA), where DNA fragment is between exon 14 and intron 14 of MDR1 gene, were selected [17]. As a forward primer and a backward primer were used respectively 5´-TGGGGCTTTTAGTGTTGGAC-3´ and 5´-TGTGGAGAGCTGGATAAAGTCA-3´. PCR amplification and melting analysis were performed on the Line Gene K (Bioer, China) using FastStart Universal SYBR Green Master (ROX) (Roche, Switzerland) as recommended by the manufacturer. The cycling protocol started with one cycle of 50 °C for 2 min followed by one cycle of 95 °C for 10 min followed by 99 cycles of 95 °C for 15 s (1 °C/cycle), a touch down of 95 °C to 61.2 °C for 1 min. The melting analysis step started of 61.2 °C to target temperature of 95 °C for 15 s (0.5 °C/cycle). Fluorescence data were converted into melting peaks by the aid of fluorescence quantitative PCR detection software (Line-Gene K, Bioer, China) to determine binding efficiency of DNA exposed to docetaxel. All samples were tested in duplicate to ensure reproducibility of the melt curves.
Sequencing DNA and Statistical Analysis
PCR products were sequenced in Sequencing Centre of Department of Molecular Biology, University of Veterinary Medicine and Pharmacy in Košice, Slovak Republic. The concentration of primer was 10 pmol/ml. The template accomplished the volume of 25 ml. The volume of primer was 0.5 ml. The resulting percentage of nucleotide bases and amino acid of MDR1 gene were analyzed with the aid of BioEdit Sequence Alignment Editor.
Statistical analysis
The results are presented as the mean ± standard deviation (SD). To determine whether sample means are significantly different from each other at the 5 % significance level single factor Anova was used.
Result and Discussion
The effect of docetaxel on the generation of DNA strand alterations derived from human adenocarcinoma MCF-7 cells was evaluated using the HRM. Fig. 1 illustrates the melting curves and the derivative of fluorescence with respect to temperature (dF/dT) of DNA of control MCF-7 cells and docetaxel treated MCF-7 cells. For all isolates, the SYBR green generated highly reproducible and sensitive melt profiles. One peak (indicated as Tmc, Tm1, Tm250) were present on the melting curves offering different characteristics for control and docetaxel treated samples (Fig. 1 and Fig. 2). Biological replicates had identical melting patterns. For the same run, variability of Tm was <0.1 %, but Tm showed 2 % to 3 % variation between runs. Such variations can be due to the limited dye stability SYBR Green.
Fig. 1: Representative melting curve (A) and derivative of fluorescence with respect to the temperature (dF/dT). 
Fig. 2: Representative melting curve of DNA of control MCF-7 cells (1), and docetaxel treated MCF-7 cells (2 - 1 µM, 3 - 250 µM). 
Based on 12 measurements, one melting peak was displayed at Tmc = 73.66 ± 2.01 °C for the control MCF-7, at Tm1 = 77.62 ± 1.88 °C and Tm250 = 82.83 ± 1.73 °C for MCF-7 at 1 µM and 250 µM of docetaxel, respectively.
Using MDR1 primer pairs: forward 5′-CCCATCATTGCAATAGCAGG-3′, backward 5′-GTTCAAACTTCTGCTCCTGA-3′, it was possible to amplify fragments of genomic DNA and detect PCR products. In Fig. 3 and Fig. 4 there are the aligned nucleotide sequences of fragments of DNA of control MCF-7 cells (C_F, C_R) and docetaxel treated MCF-7 cells (1_F, 1_R - 1 µM, 5_F, 5_R - 250 µM) for forward and backward primers, respectively.
Fig. 3: The aligned nucleotide sequences of fragments of DNA of control MCF-7 cells (C_F) and docetaxel treated MCF-7 cells (1_F - 1 µM, 5_F - 250 µM) for forward primers. 
Fig. 4: The aligned nucleotide sequences of fragments of DNA of control MCF-7 cells (C_R) and docetaxel treated MCF-7 cells (1_R - 1 µM, 5_R - 250 µM) for backward primers. 
The sequencing results confirm different nucleotide sequences for both fragments of the control and docetaxel-treated DNA. Apparently, docetaxel treatment gave rise to the deletions of the bases on the forward end of the DNA chain. Frequency of deletions was increasing with the concentration of the drug. Except for deletions, there are transitions visible, e.g. on the chain C_F/1_F on the position 136 (A → T) or C_F/5_F on the position 291 (T → C). On the backward end of the DNA chain, the insertions and transitions of bases can be observed: transitions in the sample C_R/1_R on positions 330, 331 (C → G) or in C_R/5_R on the position 114 (C → A).
Table I gives the representation of the molecular profile of DNA extracted from human adenocarcinoma MCF-7/c cells and docetaxel-treated cells - MCF-7/1 (1 µM) and MCF-7/250 (250 µM). On the forward end, comparing of the relative content G+C/A+T the increase from 71.56 % in MCF-7/c to 76.09 % and 78.41 % for MCF-7/1 and MCF-7/250, respectively, was observed. Contrary to the forward primer, the relative nucleotide ratio G+C/A+T for the backward primer decreases from 88.24 % for control set to 82.12 % for docetaxel treated cells. As can be seen from Fig. 5 and Fig. 6, the alterations in amino acid composition coded by fragments of DNA from control MCF-7 cells in comparison with docetaxel treated cells were detected. The sequencing results did not confirm identical nucleotide sequences for both fragments of DNA.
Tab. 1. Tab. 1: The basic characteristics and nucleotide composition of DNA (MCF-7/c; MCF-7/1; MCF-7/250) for the forward and the backward primer. 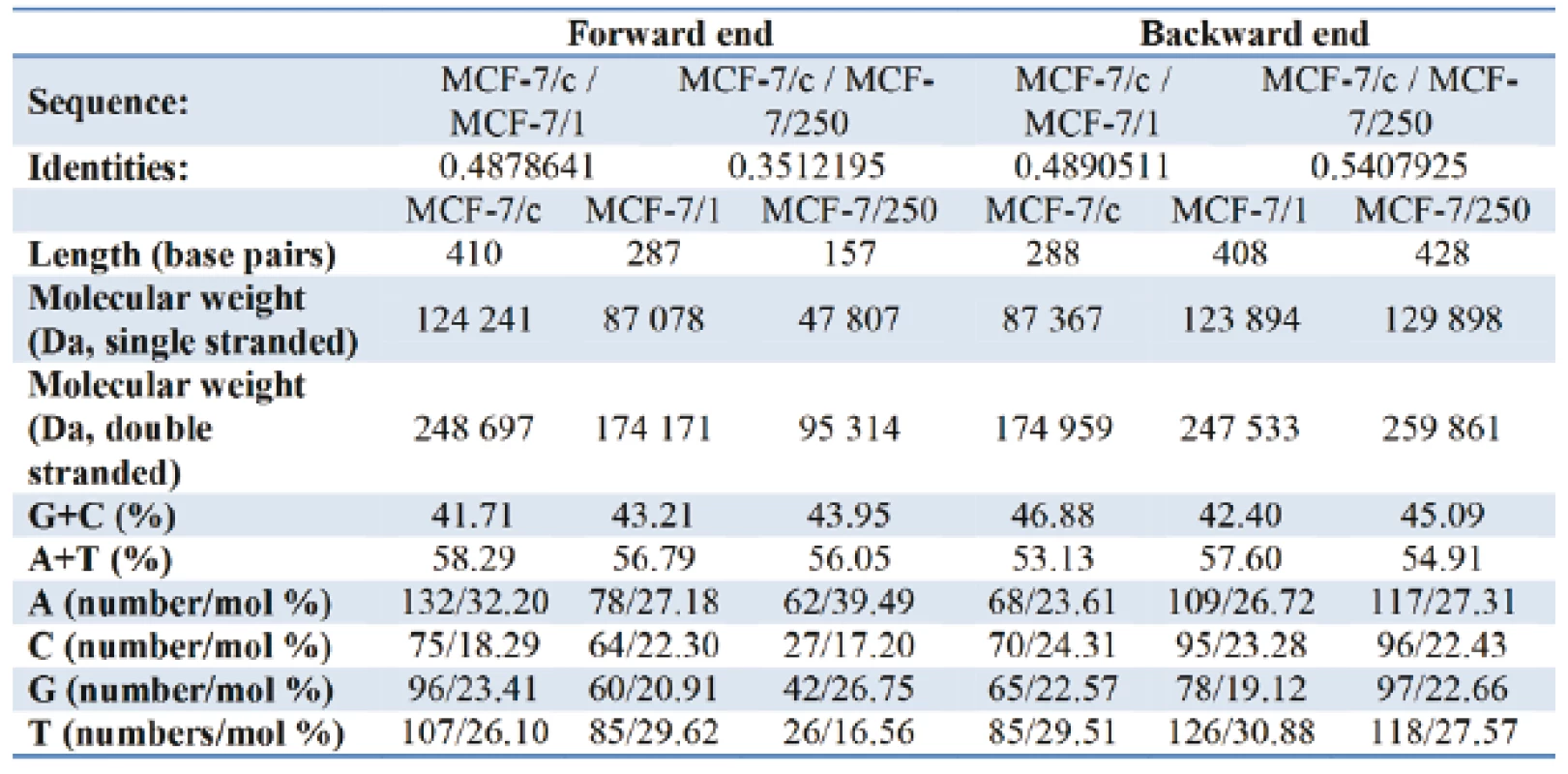
Fig. 5: Amino acid composition of DNA (MCF-7/c; MCF-7/1; MCF-7/250): the forward. 
Fig. 6: Amino acid composition of DNA (MCF-7/c; MCF-7/1; MCF-7/250): and the backward end. 
Conclusion
The melting temperature of DNA, that is defined as the temperature at which half of the DNA double helix dissociates into single strands, is related, in general, to the stability and the specific nucleotide sequence of the macromolecule. The melting curves of DNA of MCF-7 cells in the absence and presence of docetaxel represented in Fig. 1 revealed a trend for increasing of Tm of DNA after treating the adenocarcinoma cells with docetaxel, suggesting that under the influence of the taxane DNA helix was stabilized.
It is well-known that the efflux of the various cytotoxic anticancer drugs is mediated by MDR1 which affects the intracellular contents and intra-tumor distribution of compounds. Using MDR1 primer pairs, our sequencing results confirmed that the adenocarcinoma MCF-7 cells are susceptible to genomic DNA instability when exposed to chemotherapy treatments containing docetaxel. The accumulation of irregularities, such as insertions and deletions of the bases may affect critical genes involved in controlling both the intracellular contents and intra-tumor distribution of the drug and cell survival and thus guide the process of multiplex steps in the progression of breast cancer. In this context, understanding the problem of the transport out of the tumor cell by drug efflux transporters is essential when investigating combination therapies of drugs that are MDR1 substrates.
Acknowledgement
This study was supported partially by the grants VEGA 1/1109/11 (50 %), VEGA 1/0515/13 of the Ministry of Education of the Slovak Republic and by the Agency of the Slovak Ministry of Education.
Jozef Živčák, Dr. h. c. prof, Ing., Ph.D.
Department of Biomedical Engineering and Measurement
Faculty of Mechanical Engineering
Technical University of Košice
Letná 9, 042 00 Košice
E-mail: jozef.zivcak@tuke.sk
Phone: +421 55 602 2381
Zdroje
[1] Vriens, B. E. P. J., Lobbezoo, D. J. A., de Hoon, J. P. J., Veeck, J., Voogd, A. C., Tjan-Heijnen, V. C. G.: If there is no overall survival benefit in metastatic breast cancer: Does it imply lack of efficacy? Taxanes as an example. Cancer Treatment Reviews 39, 2013, 189–198.
[2] Kim, S. J., Noguchi, S.: Prediction of response to docetaxel in breast cancer. Gan To Kagaku Ryoho 35, 2008, 190–193.
[3] Ringel, I., Horwitz, S. B.: Studies with RP 56976 (taxotere): a semisynthetic analogue of taxol, J Natl Cancer Inst 83, 1991, 288–291.
[4] De Hoon, J. P. J., Veeck, J., Vriens, B .E. P. J., Calon, T. G. A., van Engeland, M., Tjan-Heijnen, V. C. G.: Taxane resistance in breast cancer: A closed HER2 circuit? Biochimica et Biophysica Acta 1825, 2012, 197–206.
[5] Torres, K., Horwitz, S. B.: Mechanisms of Taxol-induced cell death are concentration dependent. Cancer Res 58, 1998, 3620–3626.
[6] Moos, P. J., Fitzpatrick, F. A.: Taxanes propagate apoptosis via two cell populations with distinctive cytological and molecular traits. Cell Growth Differ 9, 1998, 687–697.
[7] Ieiri, I., Takane, H., Otsubo, K.: The MDR1 (ABCB1) gene polymorphism and its clinical implications. Clin Pharmacokinet 43, 2004, 553–576.
[8] Cao, D-X., Qiao, B., Ge, Z-Q., Yuan, Y-J.: Comparison of burst of reactive oxygen species and activation of caspase-3 in apoptosis of K562 and HL-60 cells induced by docetaxel. Cancer Letters 214, 2004, 103–113.
[9] Iida, S., Shimada, J., Sakagami, H.: Cytotoxicity induced by docetaxel in human oral squamous cell carcinoma cell lines in vivo. 27, 2013, 321–332.
[10] Saleh, E. M., El-awady, R. A., Anis, N., El-sharkawy, N.: Induction and repair of DNA double-strand breaks using constant-field gel electrophoresis and apoptosis as predictive markers for sensitivity of cancer cells to cisplatin. Biomedicine & Pharmacotherapy 66, 2012, 554-562.
[11] Gonzalez-Angulo, A. M., Morales-Vasquez, F., Hortobagyi, G. N.: Overview of resistance to systemic therapy in patients with breast cancer. Adv Exp Med Biol, 2007, 608 : 1–22.
[12] He, S., Liu, F., Xie, Z., Zu, X., Xu, W., Jiang, Y.: P-Glycoprotein/MDR1 regulates pokemon gene transcription through p53 expression in human breast cancer cells. Int J Mol Sci 11, 2010, 3309-051.
[13] Zhang, F., Zhang, H., Wang, Z., Yu, M., Tian, R., Ji, W., Yang, Y., Niu, R.., P. Glycoprotein associates with Anxa2 and promotes invasion in multidrug resistant breast cancer cells. Biochemical Pharmacology 2013.
[14] Takano, E. A., Mitchell, G., Fox, S. B., Dobrovic, A.: Rapid detection of carriers with BRCA1 and BRCA2 mutations using high resolution melting analysis. BMC Cancer 59, 8, 2008.
[15] Reed, G. H., Wittwer, C. T.: Sensitivity and specificity of SNP scanning by high-resolution melting analysis. Clin Chem 50, 2004, 1748-1754.
[16] Trebuňová, M., Laputková, G., Lacjaková, K., Verebová, A., Géci, I., Sabo, J.: Molecular melting profile of MDR1 gene in doxorubicin and docetaxel treated MCF-7 and natural MCF-7 cell line. European Journal of Experimental Biology 2, 2012, 449-453.
[17] Sung-Tsai, Y., Tzer-Ming, Ch., Shih-Yun, T., Yen-Hui, Ch.: Tryptanthrin inhibits MDR1 and reverses doxorubicin resistance in breast cancer cells. Biochem Biophys Res Commun 358, 2007, 79-84.
Štítky
Biomedicína
Článok vyšiel v časopiseLékař a technika

2014 Číslo 1-
Všetky články tohto čísla
- Rational operation of MRI equipment in university hospitals in the Czech Republic
- Individualization of head related transfer function
- Gene expression profiling after angiogenesis inhibitor treatment
- The effect of acetylsalicylic acid on angiogenesis in vitro
- The effect of docetaxel on molecular melting profile of DNA extracted from human breast adenocarcinoma MCF-7 cells
- Metallic nanoparticles affected by therapeutic ultrasound – The in vitro study of cell viability
- A method of compliance measurement and gastight testing in models of the respiratory system
- Lékař a technika
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- A method of compliance measurement and gastight testing in models of the respiratory system
- Rational operation of MRI equipment in university hospitals in the Czech Republic
- Metallic nanoparticles affected by therapeutic ultrasound – The in vitro study of cell viability
- Individualization of head related transfer function
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



