-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
The Dot/Icm System Delivers a Unique Repertoire of Type IV Effectors into Host Cells and Is Required for Intracellular Replication
Coxiella burnetii, the causative agent of human Q fever, is an intracellular pathogen that replicates in an acidified vacuole derived from the host lysosomal network. This pathogen encodes a Dot/Icm type IV secretion system that delivers bacterial proteins called effectors to the host cytosol. To identify new effector proteins, the functionally analogous Legionella pneumophila Dot/Icm system was used in a genetic screen to identify fragments of C. burnetii genomic DNA that when fused to an adenylate cyclase reporter were capable of directing Dot/Icm-dependent translocation of the fusion protein into mammalian host cells. This screen identified Dot/Icm effectors that were proteins unique to C. burnetii, having no overall sequence homology with L. pneumophila Dot/Icm effectors. A comparison of C. burnetii genome sequences from different isolates revealed diversity in the size and distribution of the genes encoding many of these effectors. Studies examining the localization and function of effectors in eukaryotic cells provided evidence that several of these proteins have an affinity for specific host organelles and can disrupt cellular functions. The identification of a transposon insertion mutation that disrupts the dot/icm locus was used to validate that this apparatus was essential for translocation of effectors. Importantly, this C. burnetii Dot/Icm-deficient mutant was found to be defective for intracellular replication. Thus, these data indicate that C. burnetii encodes a unique subset of bacterial effector proteins translocated into host cells by the Dot/Icm apparatus, and that the cumulative activities exerted by these effectors enables C. burnetii to successfully establish a niche inside mammalian cells that supports intracellular replication.
Published in the journal: The Dot/Icm System Delivers a Unique Repertoire of Type IV Effectors into Host Cells and Is Required for Intracellular Replication. PLoS Pathog 7(5): e32767. doi:10.1371/journal.ppat.1002056
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002056Summary
Coxiella burnetii, the causative agent of human Q fever, is an intracellular pathogen that replicates in an acidified vacuole derived from the host lysosomal network. This pathogen encodes a Dot/Icm type IV secretion system that delivers bacterial proteins called effectors to the host cytosol. To identify new effector proteins, the functionally analogous Legionella pneumophila Dot/Icm system was used in a genetic screen to identify fragments of C. burnetii genomic DNA that when fused to an adenylate cyclase reporter were capable of directing Dot/Icm-dependent translocation of the fusion protein into mammalian host cells. This screen identified Dot/Icm effectors that were proteins unique to C. burnetii, having no overall sequence homology with L. pneumophila Dot/Icm effectors. A comparison of C. burnetii genome sequences from different isolates revealed diversity in the size and distribution of the genes encoding many of these effectors. Studies examining the localization and function of effectors in eukaryotic cells provided evidence that several of these proteins have an affinity for specific host organelles and can disrupt cellular functions. The identification of a transposon insertion mutation that disrupts the dot/icm locus was used to validate that this apparatus was essential for translocation of effectors. Importantly, this C. burnetii Dot/Icm-deficient mutant was found to be defective for intracellular replication. Thus, these data indicate that C. burnetii encodes a unique subset of bacterial effector proteins translocated into host cells by the Dot/Icm apparatus, and that the cumulative activities exerted by these effectors enables C. burnetii to successfully establish a niche inside mammalian cells that supports intracellular replication.
Introduction
Coxiella burnetii, a Gram-negative facultative intracellular bacterium, is the causative agent of human Q fever, a worldwide zoonosis predominantly linked to exposure to domesticated livestock. Q fever typically manifests as an acute flu-like illness, however, chronic Q fever can also develop, typically in immunocompromised individuals [1].
Fundamental to the pathogenesis of C. burnetii is the capacity to replicate within a specialized vacuole that is derived from host lysosomes. C. burnetii requires the low pH environment of the lysosome to convert from the environmentally-resistant small cell variant that is the infectious form of the bacterium to a large cell variant that represents the replicative form of the bacterium [2], [3]. The Coxiella-containing vacuole (CCV) retains the degradative capacity of a lysosome [4], and is decorated by the host lipid raft proteins flotilin 1 and 2 and the autophagosome marker LC3 [5], [6]. The CCV also undergoes expansion, often occupying the majority of the host cytoplasm, through robust homotypic fusion with endolysosomal vesicles [7]. Importantly, the formation and development of this fusogenic CCV is dependent on bacterial protein synthesis [7], implying that C. burnetii directs formation of the vacuole in which it resides.
C. burnetii proteins important for establishment of this specialized vacuole are predicted to be translocated into the cytosol of the host cell by the Dot/Icm type IV secretion system. This secretion system has both sequence homology and functional similarity to the Dot/Icm apparatus of Legionella pneumophila [8]–[10], which is involved in manipulating cellular functions in the protozoan hosts that L. pneumophila has coevolved with in nature [11], and also in mammalian cells that represent accidental hosts for this bacterium [12]–[14]. Within these evolutionarily diverse phagocytic host cells, the L. pneumophila Dot/Icm system is essential for establishment of a unique endoplasmic reticulum-derived vacuole that enables intracellular survival and replication of this pathogen [15]–[17]. It is estimated that L. pneumophila is capable of translocating over 200 different proteins using the Dot/Icm system [18], [19]. Loss of a single effector protein in L. pneumophila does not typically diminish intracellular replication, indicating a degree of functional redundancy among the effectors that is not resolved through standard approaches involving forward genetic analysis. Defined aspects of the L. pneumophila vacuole morphology, however, can be linked to specific Dot/Icm effector proteins. For example, the effector DrrA (SidM) has a specific role in manipulating the function of the host GTPase Rab1 and promoting the localization of Rab1 to vacuoles containing L. pneumophila [20]–[22]. Similarly, the effector RalF recruits the host GTPase Arf1 to vacuoles, and ralF mutant bacteria occupy vacuoles that fail to recruit Arf1 to their limiting membrane [23].
Although proteins that have limited regions of homology with L. pneumophila effectors are encoded by C. burnetii, this intracellular pathogen does not appear to produce bone fide homologues of L. pneumophila Dot/Icm effector proteins. This suggests that these pathogens possess unique effector repertoires that could reflect the divergent pathways that have resulted in these organisms occupying unique replicative niches within evolutionarily diverse eukaryotic hosts. L. pneumophila was used previously as a surrogate to demonstrate several C. burnetii Ank proteins containing ankyrin repeat homology domains are translocated into mammalian hosts by a Dot/Icm-dependent mechanism [24], [25]. Comparative genomics revealed a high degree of variation of these Ank proteins among different isolates of C. burnetii, with mutations rendering many Ank coding regions as putative pseudogenes [25]. One of the few Ank proteins conserved among the sequenced C. burnetii isolates is AnkG, which has been demonstrated to mediate potent anti-apoptotic activity when translocated into mammalian host cells by the Dot/Icm system [26]. Given the diverse repertoire of Dot/Icm effectors encoded by L. pneumophila, there should be many novel effectors encoded by C. burnetii in addition to the Ank proteins.
The goal of this study was to identify new effectors from C. burnetii by conducting an unbiased screen for type IV secretion signals capable of delivering the calmodulin-dependent adenylate cyclase protein into host cells by the Dot/Icm system, and determine whether the delivery of effectors by C. burnetii is important for host cell infection.
Results
A screen for type IV translocation signals identifies novel C. burnetii effectors
Most Dot/Icm effectors contain a translocation signal recognized by the type IV apparatus, which is typically located near the carboxyl terminal region of the protein [18], [25], [27]. Thus, an unbiased genetic screen was designed to identify C. burnetii proteins having Dot/Icm-dependent translocation signals. A library was constructed by inserting random fragments of genomic DNA from C. burnetii RSA493 Nine Mile phase II (NM) downstream of a plasmid-encoded adenylate cyclase (Cya) enzyme that would serve as a reporter for translocation of proteins into the cytosol of host cells by the Dot/Icm system. The C. burnetii genomic DNA fragments were generated by limited digestion with the restriction enzyme SauIIIA1 and ligation of the resulting DNA fragments downstream of a cya reporter in the plasmid pEC33. The resulting plasmid library was introduced into L. pneumophila to screen for C. burnetii genes containing a type IV translocation signal that when fused in frame with cya would result in a hybrid protein delivered into mammalian host cells by the Dot/Icm system.
Because the possibility of a single plasmid in the library having a DNA fragment inserted in the correct orientation and in frame with cya was predicted to be roughly one in six, the decision was made to screen a pool of bacteria containing different plasmid clones rather than initially screening individual clones from the library. To determine the feasibility of identifying a single translocated effector in pools of transformants, L. pneumophila expressing Cya-RalF, a fusion protein known to be efficiently translocated by the Dot/Icm system [23], [27], and L. pneumophila producing Cya alone, were mixed at defined ratios. Infection at a ratio of bacteria producing Cya-RalF to bacteria producing the control Cya of 1∶50 reproducibly led to an increase in host cell cAMP levels of at least 2.5-fold over background levels (Figure S1). These data indicated that it should be possible to detect a single positive clone with a type IV-dependent translocation signal fused to Cya in a mixed pool containing roughly 50 negative clones. Thus, the library of clones was distributed into pools that contained an estimated 50 different transformants harboring a randomly inserted C. burnetii gene downstream of cya (Figure S2). Mammalian CHO FcγRII cells [28] were infected with the pools of L. pneumophila containing Cya fused randomly to C. burnetii gene products. Screening identified pools that resulted in a significant increase in host cAMP levels above background, and these pools were further analyzed to isolate individual clones with an in-frame fusion between cya and a C. burnetii gene having a functional Dot/Icm translocation signal.
A total of 22-positive clones were identified from the 506 different pools tested in the translocation assay, which would theoretically represent the screening of 4200 in-frame fusions. Plasmids were isolated from the positive clones and DNA sequencing was used to identify the predicted C. burnetii protein fused to Cya. This analysis resulted in the identification of 11 potential Dot/Icm effector proteins (Table S1), including the previously characterized Dot/Icm effector AnkG (CBU0781) [24]–[26].
To validate that the proteins identified in the screen were bone fide type IV effectors, the predicted full-length open reading frames for each of the 10 new proteins identified in the screen were amplified using genomic DNA from the C. burnetii NM strain, and the resulting full-length NM gene was fused to the C-terminus of Cya. Plasmids encoding a putative full-length effector fused to Cya were introduced into a strain of L. pneumophila having a functional Dot/Icm system and a ΔdotA mutant strain with a non-functional type IV apparatus [15]. Seven of the 10 C. burnetii fusion proteins showed significant increases in cAMP levels compared to controls (Figure 1A). Importantly, translocation was observed to be dependent upon a functional Dot/Icm system as cAMP levels were unchanged after infection with the ΔdotA strain (Figure 1A). The three fusion proteins that did not demonstrate translocation; Cya-1780, Cya-1957 and Cya-2064 were still expressed at levels comparable to Cya and Cya-RalF in L. pneumophila (Figure S3).
Fig. 1. Dot/Icm-dependent translocation of C. burnetii proteins by L. pneumophila. 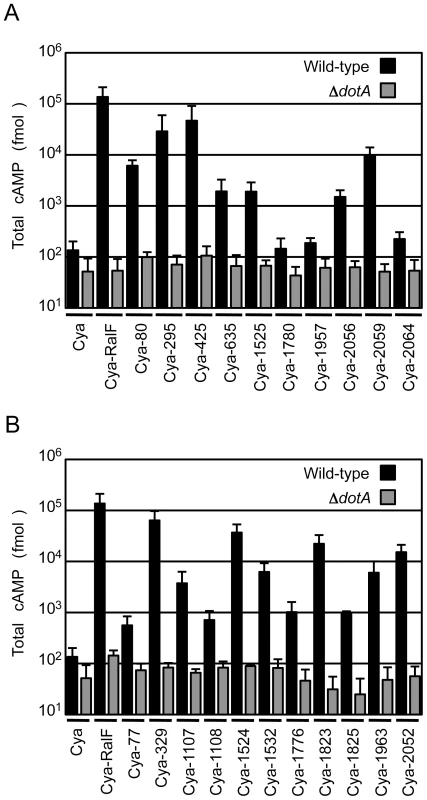
CHO-FcγRII cells were infected with Dot/Icm-sufficient LP01 strain of L. pneumophila (black) or the isogenic ΔdotA mutant (grey) expressing Cya fusions to the indicated C. burnetii proteins. (A) Fusions to Cya of the indicated full-length derivatives of C. burnetii NM proteins identified in the genetic screen were tested for translocation (B). Fusions to Cya of the indicated full-length derivatives of C. burnetii NM proteins identified based on homology or proximity to proteins identified in the screen were tested for translocation. Cya indicates empty vector control. Cya-RalF was used as a positive control. After 1 h, host cells were lysed and cAMP was extracted. Total cAMP levels resulting from translocation of protein fusions were quantified using an enzyme-immunoassay system, and are shown in fmol. Results represent average values +/− SD of experiments performed in triplicate. Because genes encoding L. pneumophila effectors are often found in clusters at distinct chromosomal locations and can be duplicated to generate families of paralogous effectors [29], we investigated whether any of the C. burnetii genes in close proximity or with extensive sequence similarity to the identified effectors might also encode effectors. Thirty-seven additional C. burnetii genes that fit these criteria were tested for translocation by L. pneumophila (Table S2). Of the 37 genes tested, there were 11 genes encoding proteins translocated into host cells by a Dot/Icm-dependent mechanism (Figure 1B). Thus, this screen in total resulted in the identification of 18 different C. burnetii proteins that are effectors delivered into host cells by the L. pneumophila Dot/Icm apparatus.
Genomic comparisons indicate plasticity among novel C. burnetii effectors
Sequence analysis revealed that none of the 18 C. burnetii Dot/Icm effectors have significant homology to proteins found in other organisms, demonstrating the unique nature of these proteins. CBU0077 from the strain NM RSA493 is highly conserved in the two different sequenced C. burnetii strains isolated from chronic Q-fever patients presenting with endocarditis (G Q212 and K Q154) and in the Dugway strain isolated from rodents [30], which does not appear to cause clinical disease. For the remaining effectors, there were polymorphisms in the coding regions that would indicate that these effectors have either been mutated or have not been acquired by one or more of the strains of C. burnetii. An alignment of these effectors in all sequenced strains is presented in Figure 2. A comparison of the NM genome to the genomes of the G, K and Dugway strains revealed nucleotide deletions or nonsense mutations in the NM genes that would predict the production of a shorter version of several effector proteins compared to homologues found in some of these other sequenced strains [31]. Interestingly, three of the NM effectors identified here, CBU1108, CBU1107 and CBU1776, are not present in the G Q212 strain (Figure 2). Thus, similar to Dot/Icm effectors in L. pneumophila, there is significant plasticity observed in the repertoire of effectors of C. burnetii when genomes from different isolates are compared [32]. Despite these polymorphisms, all of the genes encoding the NM Dot/Icm effectors identified here were transcribed during infection (Figure S4). Thus, the genes encoding truncated NM effectors are transcribed and the predicted translated products should be delivered into host cells during infection by the functional Dot/Icm translocation signal identified in the protein.
Fig. 2. Domain analysis and genomic comparisons for C. burnetii effectors identified in this study. 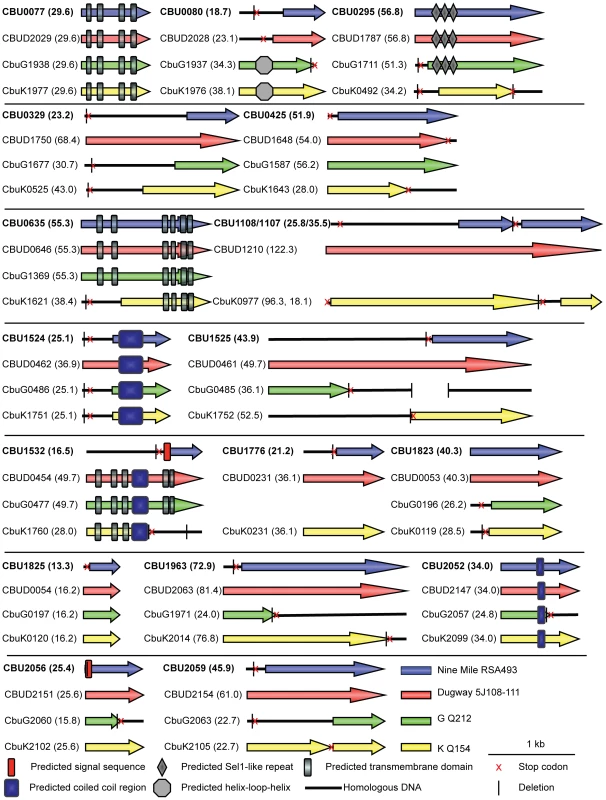
The schematic shows the eighteen NM effectors identified in this study represented in blue. Below each NM effector is a representation of the size of the homologous reading frame encoded by the genes in the other sequenced strains of C. burnetii. The size of the predicted proteins is represented in kDa in brackets following the gene designation. Black lines represent the presence of homologous DNA that does not encode an open reading frame due to small deletions (vertical black line) and stop codons (red cross). In cases where multiple deletions occur the first deletion, representing the site of the frameshift, is displayed. The locations of conserved domains in each protein identified in a SMART database search are indicated according to the key provided. Structural features and host cell localization phenotypes exhibited by C. burnetii effectors
Analysis of the C. burnetii effectors using the Simple Modular Architecture Research Tool (SMART) database [33] revealed domains in these proteins that could provide insight into potential functions (Figure 2). Identified features included predicted transmembrane domains (CBU0077/CBUD2029/CbuG1938/CbuK1977, CBU0635/CBUD0646/CbuG1369/CbuK1621, and CBUD0454/CbuG0477/CbuK1760), proteins with predicted coiled-coil regions (CBU1524/CBUD0462/CbuG0486/CbuK1751, CBUD0454/CbuG0477/CbuK1760, and CBU2052/CBUD2147/CbuG2057/CbuK2099), an effector with three Sel1-like repeats (SLRs; CBU0295/CBUD1787/CbuG1711) and a protein with a helix-loop-helix motif (CbuG1937/CbuK1976). SLR-containing proteins, found in eukaryotes and bacteria, are thought to mediate protein-protein interactions and have been found in several pathogenic bacteria. Of note, two of the L. pneumophila SLR-containing proteins, EnhC and LpnE, have been associated with virulence [34]–[36].
To further characterize the identified Dot/Icm effectors, the products encoded by the NM genes and the longer derivatives encoded by the Dugway, G or K strains, were ectopically produced in HeLa 229 cells as proteins containing three tandem copies of the N-terminal FLAG epitope tag. All of the NM effectors demonstrated similar localization profiles as the homologous proteins isolated from the other C. burnetii strains, with the exception of CBU0080. The NM CBU0080 protein showed diffuse cytosolic distribution compared to the CbuK1976 protein, which localized to the nucleus (Figure 3A). The N-terminal 19.4 kDa region in CbuK1976 that is absent in CBU0080 encodes a predicted nuclear localization signal (NLS) containing the peptide KKRK in addition to a helix-loop-helix motif.
Fig. 3. Effectors of C. burnetii show differential localization patterns when expressed in mammalian HeLa 229 cells. 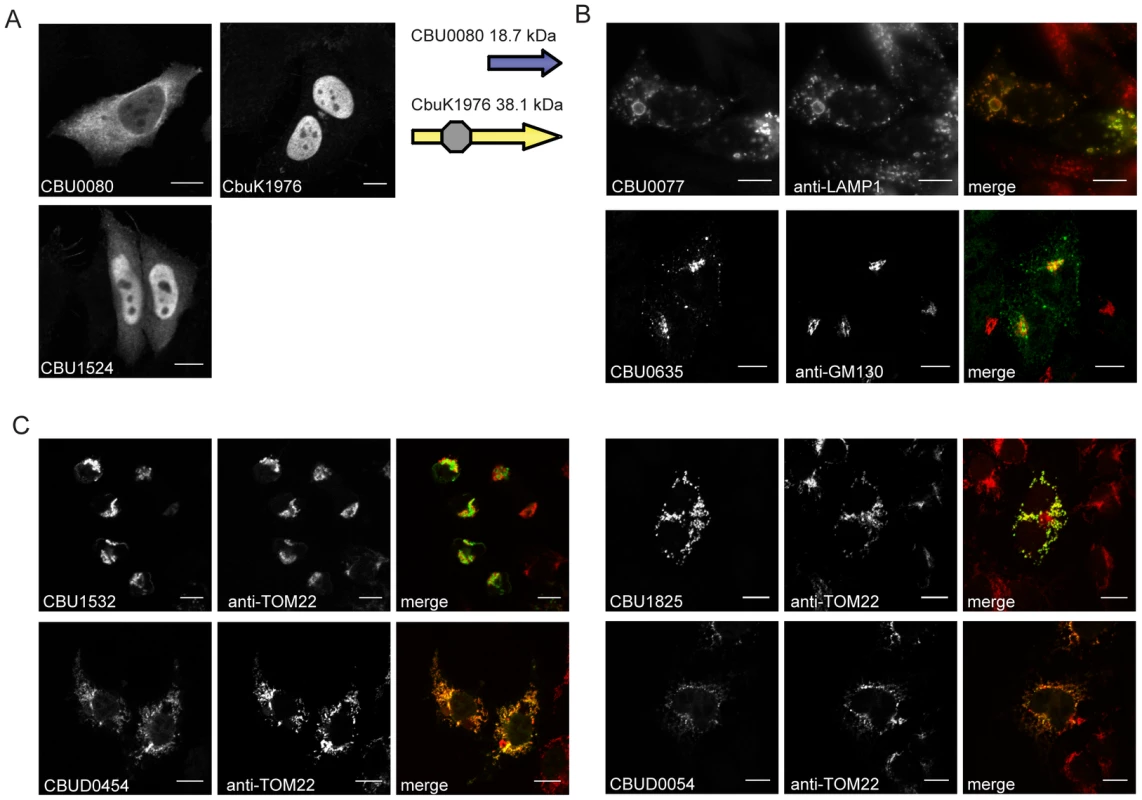
Immunofluorescence micrographs show the representative localization profiles of the indicated FLAG-tagged C. burnetii effectors (Green) and the indicated host organelles (Red) after ectopic production of the proteins in HeLa 229 cells. (A) CbuK1976 and CBU1524 show localization to the nucleus, but the NM protein CBU0080 is distributed throughout the cytoplasm. The schematic shows that CBU0080 does not contain the helix-loop-helix motif (octagon) found in the homolog CbuK1976, which could explain the differential localization phenotypes observed (B) Cells stained for FLAG-tagged CBU0077 and LAMP1 show colocalization of the effector on lysosome-derived organelles, and cells stained for CBU0635 and GM130 show distribution of the protein to a perinuclear region containing the Golgi apparatus. (C). Cells stained for FLAG-tagged effectors and the TOM22 protein show colocalization of the effectors on mitochondria. The NM protein CBU1825 and the homologous Dugway protein CBUD0054 showed similar localization patterns. The NM protein CBU1532 and the Dugway homolog CBUD0454 showed slightly different mitochondrial straining patterns, with CBU1532 production leading to cell rounding and mitochondria aggregation. Scale bars represent 10 µm. Most of the proteins localized to the host cell cytosol or diffusely throughout the cell when expressed in HeLa 229 cells (Table 1 and Figure S5). Several proteins showed enrichment in specific subcellular compartments as determined by colocalization with proteins that label these host organelles. Similar to CbuK1976, the CBU1524 protein localized to the nucleus when produced in mammalian cells (Figure 3A). Nuclear localization of CBU1524 may be mediated by a predicted seven residue NLS containing the peptide PKRTRVD that begins at amino acid position 182 in the protein. The protein CBU0077 resided primarily on lysosomes as demonstrated by colocalization with lysosomal-associated membrane protein 1 (LAMP1). The protein CBU0635 was present at a perinuclear position juxtaposed to the Golgi apparatus (Figure 3B). Two NM effector proteins, CBU1532 and CBU1825, along with the respective homologues from Dugway, CBUD0454 and CBUD0054, showed a specific affinity for mitochondria (Figure 3C). No defined mitochondrial targeting sequences were apparent in any of these effector proteins. Interestingly, CBU1532 appeared to affect the morphology of HeLa 229 cells, with many transfected cells having a rounded appearance with aggregated mitochondria. The Dugway protein, CBUD0454, did not mediate this morphological change when expressed in HeLa 229 cells, which might indicate that CBU1532 confers a dominant-negative effect that perturbs cellular functions modulated by CBUD0454. Similar localization patterns were observed in CHO-FcγRII cells and HEK 293 cells (data not shown), indicating that protein localization patterns are not cell-type specific.
Tab. 1. Characteristics of C. burnetii effector proteins. 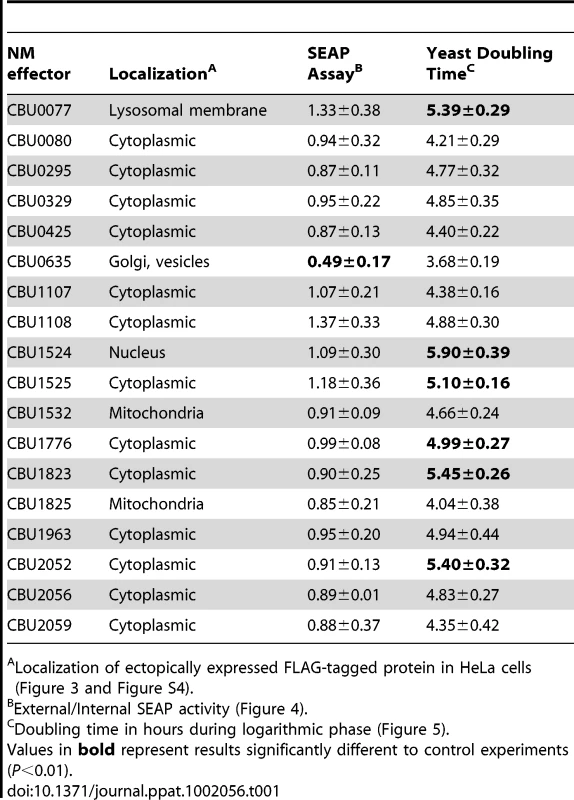
Localization of ectopically expressed FLAG-tagged protein in HeLa cells (Figure 3 and Figure S4). CBU0635 overproduction interferes with the host secretory pathway
Several L. pneumophila Dot/Icm effectors have been shown to disrupt vesicle trafficking in mammalian cells by affecting the dynamics of membrane transport in the early secretory pathway [20], [21], [23], [24]. To investigate if any of the identified C. burnetii proteins have effector functions that interfere with the secretory pathway in mammalian cells, transport of secreted alkaline phosphatase (SEAP) was investigated in HEK 293 cells ectopically expressing the 3×FLAG-tagged effector proteins. The amount of SEAP secreted by cells into the culture medium was compared to the amount of SEAP that remained intracellular after an 8 h incubation period (Table 1 and Figure 4A). The negative control was HEK 293 cells transfected with pFLAG alone, showing an external/internal SEAP activity ratio of 1.23±0.46. The positive control was HEK 293 cells transfected with a plasmid with the GDP-locked allele ARF1T31N, showing a SEAP activity ratio of 0.60±0.21. Expression of the C. burnetii Dot/Icm effector CBU0635 was found to interfere with the secretory pathway based on an external/internal SEAP activity ratio of 0.49±0.17. Consistent with CBU0635 being the only effector that showed a pattern of localization near the Golgi apparatus when ectopically expressed in eukaryotic cells (Figure 3B), it was the only effector found to interfere with the host secretory pathway. Time course measurements determined that CBU0635-expressing cells had a defect in the kinetics of SEAP secretion that was similar to cells producing ARF1T31N (Figure 4B). Thus, CBU0635 has an effector activity that when overproduced disrupts the mammalian secretory pathway.
Fig. 4. CBU0635 interferes with host protein secretion. 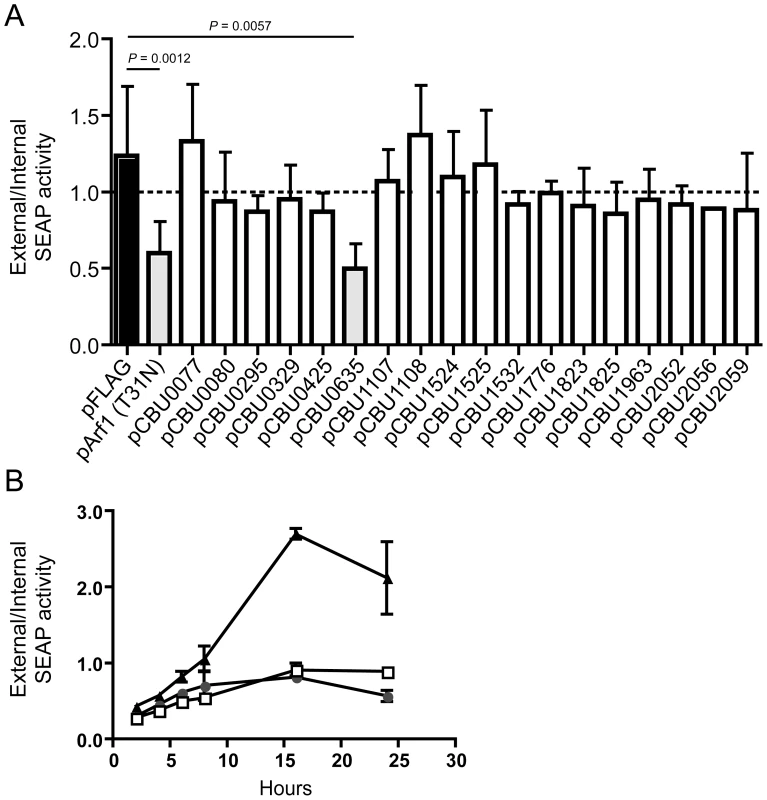
The impact of C. burnetii Dot/Icm effectors on the host cell secretory pathway was examined by monitoring secretion of the SEAP protein into the culture supernatant. (A) HEK 293 cells were co-transfected with pSEAP and the plasmid encoding the indicated effector protein or the GDP-locked ARF1T31N protein (pArf1-T31N) or empty vector (pFLAG). External and internal SEAP activity was measured. The grey bars show that there was a significant decrease in the external/internal ratio of SEAP activity observed upon ectopic expression of ARF1T31N (P = 0.0012) or CBU0635 (P = 0.0057) in comparison to HEK 293 cells transfected with pFLAG alone (black bar). Expression of all other C. burnetii effectors did not significantly the ratio of SEAP activity (white bars). (B). SEAP activity (y-axis) was measured at the indicated times after cells were washed and new culture medium was added (x-axis) Data show SEAP ratios for cells with vector alone (pFLAG; black triangles), cells producing CBU0635 (pCBU0635; open squares) and cells producing ARF1T31N (grey circles). A similar defect in SEAP secretion was observed in cells producing ARF1T31N as in cells producing CBU0635. Several C. burnetii Dot/Icm effectors slow yeast replication when overproduced
Several studies have demonstrated that expression of L. pneumophila effector proteins in Saccharomyces cerevisiae can interfere with the rate of yeast replication [37]–[39]. The L. pneumophila effector RalF interferes with yeast replication by virtue of it being able to function as a guanine nucleotide exchange factor for ARF [37]. Inhibition of yeast replication was the basis for identification of the L. pneumophila Dot/Icm effectors YlfA and YlfB [37]. To test whether the C. burnetii Dot/Icm effectors had similar activities, these proteins were produced in S. cerevisiae and growth was monitored in liquid YNB supplemented with 2% galactose to induce effector expression. None of the NM Dot/Icm effectors conferred a yeast growth phenotype equivalent to that seen with expression of YlfA (Figure 5A). To determine yeast growth rates, growth curves were performed in duplicate wells in at least three independent experiments. The doubling time of each yeast strain was calculated during the exponential phase of growth between 14 h and 18 h (Figure 5B and Table 1). The negative control strain consisting of S. cerevisiae pYES2 had a doubling time of 4.4±0.18 h. Expression of YlfA increased the doubling time to 30.94±18.74 h, P = 0.013. Six C. burnetii effectors significantly altered the growth rate of S. cerevisiae; CBU0077 5.39±0.29 h (P = 1.9×10−4), CBU1524 5.90±0.39 h (P = 5.0×10−5), CBU1525 5.10±0.16 h (P = 5.2×10−4), CBU1776 4.99±0.27 h (P = 0.0054), CBU1823 5.46±0.29 h (P = 1.7×10−4) and CBU2052 5.40±0.32 h (P = 5.5×10−4). Thus, expression of these six C. burnetii proteins moderately slows yeast replication.
Fig. 5. Several C. burnetii effectors slow yeast replication. 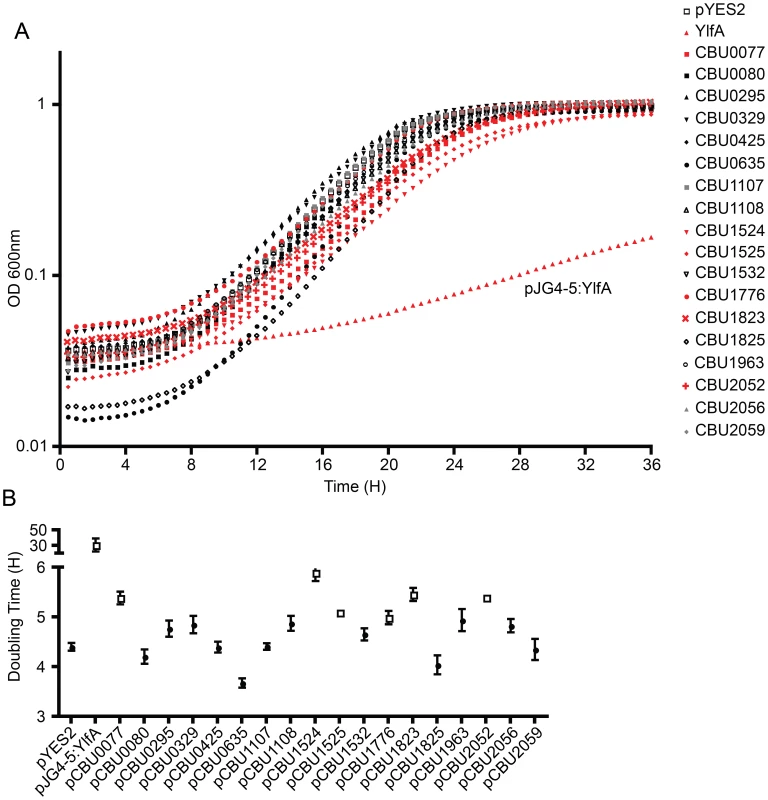
(A) Yeast strains were grown in YNB supplemented with 2% galactose to induce the expression of the indicated effector proteins listed in the legend on the right. The plots show yeast replication as determined by measuring the optical density of the culture at 600 nm (OD 600 nm, y-axis) every 30 min (x-axis) for a period of 36 h. Growth of S. cerevisiae expressing C. burnetii effectors were compared to controls that included S. cerevisiae containing the vector control (pYES2; open squares) and S. cerevisiae producing the L. pneumophila effector YlfA (pJG4-5:YlfA; red triangles). Effectors that resulted in a delay in the doubling rate of S. cerevisiae are highlighted with red symbols. (B) The doubling time of S. cerevisiae producing the indicated effector proteins was calculated during the exponential phase of growth from the growth curves shown in panel A. Effectors that resulted in a significant increase in doubling time compared to S. cerevisiae pYES2 are highlighted in open boxes (P<0.01). Data represent the mean doubling time ± SD determined from at least 3 independent growth curves. The Dot/Icm secretion system is essential for C. burnetii intracellular replication
To determine the cumulative role effector proteins may play during infection of host cells by C. burnetii, a genetic screen was initiated to identify C. burnetii having a gene essential for Dot/Icm transporter function disrupted. Towards this end a mariner-based Himar1 C9 transposase system was used to generate random insertions in the chromosome of C. burnetii NM [40]. Kanamycin-resistant transposon mutants of C. burnetii were selected on defined ACCM medium and then screened by PCR for insertions into the dot/icm locus. A mutant was isolated with the transposon inserted at base 1569472 in the chromosome, disrupting the icmL gene (also called icmL.1 and dotI) predicted to be essential for function of the Dot/Icm system (Figure 6A). Southern blot analysis and PCR amplification of the insertion site was used to confirm the site of transposon integration and to determine that there was a single copy of the transposon in this mutant (Figure 6B and C).
Fig. 6. The C. burnetii Dot/Icm system is essential for intracellular replication. 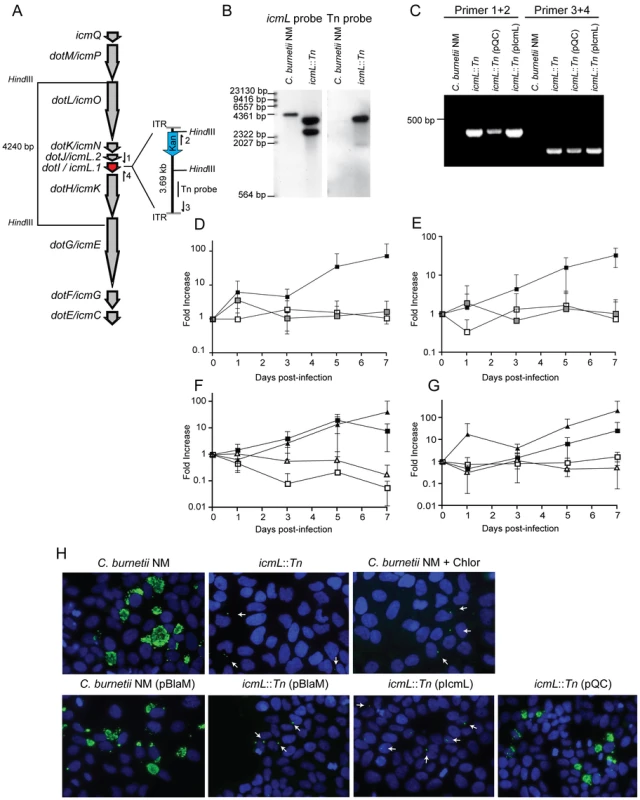
(A) A clone of C. burnetii with a 3.69 kb transposon inserted in the icmL gene (dotI/icmL.1) was isolated after enrichment on medium containing kanamycin. The schematic shows the predicted location and orientation of the transposon in icmL as determined by sequence analysis. The location of probes and restriction sites used to validate the site of transposon insertion by PCR and Southern hybridization are indicated. (B) As indicated, genomic DNA digested with HindIII isolated from the C. burnetii NM strain and the isogenic icmL::Tn mutant was analyzed by Southern hybridization using a probe derived from the icmL gene and a probe derived from the transposon. As predicted from the schematic in panel A, the icmL probe identified a single band of 4.2 kb in the NM strain, and two bands of 3.3 kb and 2.5 kb in the icmL::Tn mutant because of a new HindIII site introduced into the icmL locus by the transposon. When a probe homologous to the transposon was used (Tn probe), a single band was identified in the icmL::Tn mutant, indicating that this strain has a single insertion of the transposon in the chromosome. (C) PCR amplification of genomic DNA from NM and the icmL::Tn mutant using primer sets shown in panel A confirm the predicted location and orientation of the transposon insertion in icmL, and that the transposon is retained by the icmL::Tn strain after introduction of a plasmid encoding icmL (pIcmL) or a plasmid encoding the entire operon harboring icmL (pQC). (D,E) The ability of the icmL::Tn mutant to replicate in HeLa (D) and Vero (E) cells was determined by measuring genomic equivalents (y-axis) at the indicated times after infection (x-axis). There was a 50 to 100-fold increase over 7 days in C. burnetii NM genomic units (black squares). No significant increase in genomic units was detected for the icmL::Tn mutant (white squares) or C. burnetii NM treated with 10 µg/ml chloramphenicol (gray squares). (F,G) The ability of the icmL::Tn mutant to replicate in HeLa (F) and Vero (G) cells was determined by measuring genomic equivalents (y-axis) at the indicated times after infection (x-axis). Replication was observed for C. burnetii NM containing pBlaM (black squares) and the icmL::Tn mutant containing the plasmid pQC (black triangles). The icmL::Tn mutant containing the vector pBlaM (white squares) or for the icmL::Tn mutant containing pIcmL (white triangles). Graphs represent the mean ± SD of at least three independent experiments. (H) Fluorescence micrographs were acquired after infection of HeLa cells for 5 days by the strains of C. burnetii indicated. An anti-Coxiella antibody (green) was used to visualize intracellular bacteria and the nucleus of the host cell was visualized by DAPI staining (blue). Replicating bacteria in large vacuoles were observed for cells infected with C. burnetii NM, C. burnetii NM containing pBlaM and the icmL::Tn mutant containing pQC. Only individual bacteria were observed inside the host cells infected with the icmL::Tn mutant, C. burnetii NM in medium with chloramphenicol, icmL::Tn containing pBlaM and icmL::Tn containing pIcmL (indicated with arrows). These are representative images from at least three independent experiments. To determine whether inactivation of the Dot/Icm system would influence the ability of C. burnetii to replicate within eukaryotic host cells, both HeLa and Vero cells were infected at a multiplicity of infection (MOI) of 50 with either the parental strain C. burnetii NM phase II or the isogenic icmL::Tn mutant and bacterial replication was assayed over a period of seven days (Figure 6D and E). No replication of the icmL::Tn mutant was observed in either cell line. The isogenic C. burnetii NM phase II strain with a functional Dot/Icm system demonstrated a 100-fold increase in genome equivalents over the same period. Immunofluorescence microscopy of infected HeLa cells stained with an anti-Coxiella antibody revealed the formation of large CCVs containing replicating C. burnetii NM, and a complete absence of large vacuoles containing replicating bacteria for the isogenic icmL::Tn mutant (Figure 6H). The intracellular replication defect observed for the isogenic icmL::Tn mutant was similar to that observed for C. burnetii NM phase II cultured with host cells in medium containing chloramphenicol (Figure 6D, E and H). Importantly, the icmL::Tn mutant grew at an equivalent rate to the control strain of C. burnetii in the defined medium ACCM (Figure S6), and the intracellular replication defect was not a consequence of reduced entry, as the icmL::Tn mutant had an equivalent capacity to enter HeLa cells as the isogenic control strain (Figure S6). Furthermore, viable icmL::Tn mutant bacteria were recovered from HeLa cells at 24 h, 72 h and 120 h post-infection (Figure S6), suggesting that the Dot/Icm apparatus is not essential for survival within HeLa cells.
Complementation studies validated that the defect in replication of the icmL::Tn mutant resulted from a loss of Dot/Icm system function. The icmL gene is centrally located in a putative operon that begins with the icmQ gene and terminates after the icmC gene (Figure 6A), which would predict that polar effects resulting from the insertion of the transposon in the icmL gene would also limit the production of the downstream icmKEGC gene products. For this reason, complementation studies were conducted using two different plasmids. The plasmid pIcmL contained only the icmL gene, whereas, the plasmid pQC contained the entire operon from icmQ to icmC. The ability of these plasmids to complement the icmL::Tn mutation was determined by assaying intracellular replication of the resulting C. burnetii strains containing these plasmids in HeLa and Vero cells compared to a positive control strain, which was C. burnetii NM containing a plasmid encoding the ß-lactamase gene (pBlaM). Intracellular replication of the icmL::Tn mutant was fully restored by the plasmid pQC as indicated by the increase in genome equivalents over time (Figure 6F, G) and the appearance of vacuoles containing replicating bacteria at day 5 post-infection (Figure 6H). Thus, the intracellular growth defect observed for the icmL::Tn mutant is linked genetically to the region of the transposon insertion and is caused by inactivation of the Dot/Icm system. As predicted, the plasmid pIcmL did not complement the icmL::Tn mutation, consistent with the tranposon insertion having a polar effect on production of the downstream icm gene products. These studies demonstrate that the Dot/Icm system and the cumulative activities of the translocated effectors are essential for establishment of a vacuole that supports C. burnetii replication in mammalian host cells.
Demonstration of Dot/Icm-dependent protein translocation in C. burnetii
To validate that the effectors identified in this screen are delivered into host cells during infection by C. burnetii, a polyclonal antibody specific for the conserved C. burnetii Dot/Icm effector CBU0077 was generated (Figure 7A). HeLa cells persistently infected with C. burnetii were separated into a soluble fraction that contained host proteins and bacterial proteins secreted or translocated during infection, and a pellet fraction containing C. burnetii proteins that remain associated with intact bacterial cells and insoluble host debris. Immunoblot analysis revealed CBU0077 in the fraction of secreted/translocated bacterial proteins, whereas, the C. burnetii translation elongation factor EF-Ts, which is not a secreted/translocated protein, was detected only in the pellet fraction. These data indicate CBU0077 is produced and translocated out of the bacterial cell during infection. When de novo synthesis of bacterial proteins in the infected cells was prevented by treatment with chloramphenicol for 16 h prior to fractionation, a slight decrease in the amount of CBU0077 was found for the secreted fraction and the pellet fraction, suggesting that CBU0077 has a relatively long half-life in both bacterial and host cell compartments (Figure 7A).
Fig. 7. The C. burnetii Dot/Icm secretion system is required for effector translocation. 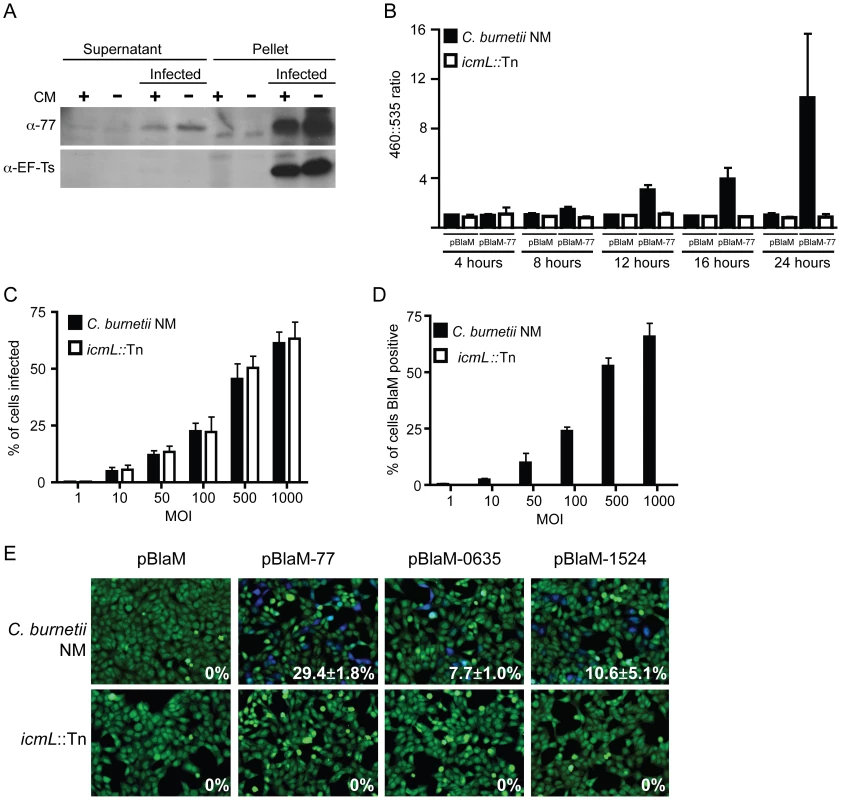
(A) HeLa cells either infected with C. burnetii NM or uninfected were incubated with or without chloramphenicol (CM) and cellular lysates were separated into supernatant and pellet fractions. Immunoblot analysis was used to detect CBU0077 (top panel; α-77) and EF-Ts (bottom panel; α-EF-Ts). The CBU0077 protein was detected in both the supernatant fraction containing proteins secreted during infection and in the pellet fraction containing intact bacteria. The cell associated EF-Ts protein was detected only in the pellet fraction containing intact bacterial cells. The addition of chloramphenicol resulted in a slight drop in levels of CBU0077 detected in the supernatant, suggesting the secreted protein has a relatively long half-life. Similar results were obtained from three independent experiments. (B) C. burnetii NM (black bars) and the icmL::Tn mutant (white bars) containing either pBlaM or pBlaM-77 were used to infect HeLa cells for the times indicated (x-axis). Translocation of the BlaM and BlaM-CBU0077 fusion protein by the respective strains was determined by measuring the change in the 460 nm/535 nm fluorescence emission ratio resulting from cleavage of the CCF4-AM substrate (y-axis). Results represent the mean ± SD obtained from triplicate samples. (C, D) HeLa cells were infected in parallel with C. burnetii NM producing BlaM-CBU0077 or the icmL::Tn strain producing BlaM-CBU0077 at the multiplicities of infection (MOI) indicated on the x-axis. Fluorescence microscopy was used to determine the percent of cells infected by the two strains (C) and the number of cells that stained positive for BlaM translocation (D) Results represent the mean ± SD obtained from triplicate samples. (E) Fluorescence micrographs of HeLa cells that were infected for 24 h with either C. burnetii NM or the icmL::Tn strains producing either BlaM alone or BlaM fusions to CBU0077 (pBlaM-77), CBU0635 (pBlaM-0635) or CBU1524 (pBlaM-1524). Emission at 535 nm (green) identifies the CCF4-AM-loaded cells and emission at 460 nm (blue) indicates cleavage of CCF4-AM in the cytosol of the infected cell resulting from translocation of the BlaM fusion protein. The percent of cells that were BlaM positive (blue) was determined for each infection condition and the mean ± SD from three independent experiments is displayed in the bottom right corner of each representative micrograph. To independently address whether CBU0077 is translocated during infection, a ß-lactamase-based translocation assay was used. In this assay, a derivative of ß-lactamase (BlaM) that lacks a signal sequence is fused to the amino terminus of a potential Dot/Icm effector. The delivery of a BlaM hybrid into the host cytosol is monitored using the substrate CCF4-AM. When BlaM fusion proteins are delivered into the host cell cytosol the CCF4-AM molecule is cleaved, resulting in a shift in fluorescence emission from 535 nm (Green) to 460 nm (Blue) when excited at 415 nm. Plasmids encoding blaM alone and the blaM-CBU0077 fusion were introduced into C. burnetii NM phase II and the isogenic icmL::Tn mutant. Production of the expected BlaM proteins was confirmed by immunoblot analysis (Figure S7). These C. burnetii strains producing BlaM proteins were used to infect HeLa cells, and cleavage of the CCF4-AM substrate was measured at 4 h, 8 h, 12 h, 16 h and 24 h post infection (Figure 7B). Translocation of BlaM and BlaM-CBU0077 was undetectable at 4 h post infection in all strains examined. At 8 h post infection, translocation of BlaM-CBU0077 was detected by C. burnetii NM phase II, with a 460 nm/535 nm fluorescence emission ratio relative to uninfected cells of 1.58±0.28 compared to 1.06±0.22 for the strain expressing BlaM alone. This signal continued to increase at 12 h (3.03±0.43), at 16 h (3.90±0.95) and at 24 h (9.17±4.96). Translocation of the BlaM protein alone by C. burnetii was not detected for any strain at any time point, confirming that delivery of BlaM into the host cytosol required fusion to the type IV effector CBU0077. Most importantly, translocation of BlaM-CBU0077 was not detected using the icmL::Tn mutant, demonstrating that the Dot/Icm apparatus is essential for delivery of this effector into the host cytosol.
Single cell assays were conducted to compare the efficiency of C. burnetii entry with the efficiency of effector translocation. Host cells were infected at different multiplicities with isogenic C. burnetii NM strains producing BlaM-CBU0077, and fluorescence microscopy was used to measure the efficiencies of infection and effector protein translocation. As the multiplicity of infection increased there was a dose-dependent increase in the number of cells that were infected by C. burnetii (Figure 7C). There was no difference observed in the infection efficiencies of the control C. burnetii NM strain compared to the isogenic icmL::Tn mutant. A dose-dependent increase in the number of cells receiving the BlaM-CBU0077 protein that closely mirrored the uptake efficiency was observed after infection with the control C. burnetii NM strain (Figure 7D), as determined by counting cells with a positive emission signal at 460 nm resulting from cleavage of the CCF4-AM substrate by the translocated fusion protein. No translocation of BlaM-CBU0077 was observed after infection of cells with the icmL::Tn mutant, even though uptake was similar to that observed for the isogenic C. burnetii NM control. These data indicate that most of the cells that are infected by C. burnetii NM are subject to Dot/Icm-dependent translocation of CBU0077.
The delivery of BlaM fusions to CBU0635 and CBU1524 was also assessed to validate that other effectors identified in this study were translocated into host cells during infection by C. burnetii by a Dot/Icm-dependent mechanism. BlaM-CBU0635 and BlaM-CBU1524 were produced in roughly equal amounts in C. burnetii NM and the isogenic icmL::Tn mutant (Figure S7). HeLa cells were scored for translocation of the BlaM fusion proteins after a 24 h infection period. Translocation-positive cells were detected after infection of host cells with C. burnetii NM producing BlaM-CBU0077, BlaM-CBU0635 and BlaM-CBU1524, however, there were no translocation-positive cells detected after host cells infection with the isogenic icmL::Tn mutant producing these fusion proteins (Figure 7E). Thus, effectors identified using the L. pneumophila system are engaged and translocated into host cells during C. burnetii infection by a Dot/Icm-dependent mechanism.
Discussion
In this study a random screen was conducted to identify novel effectors produced by C. burnetii. Because type IV secretion systems recognize translocation signals at the carboxyl terminus of effectors, a strategy was employed in which a Cya reporter that lacks a translocation signal was used to monitor the presence of coding determinants fused at the carboxyl terminus of the protein that enabled Cya to be engaged and delivered into host cells by the Dot/Icm system. This approach was successful at identifying C. burnetii proteins delivered into host cells by the Dot/Icm system, demonstrating the feasibility in using type IV translocation signal screening as a means to identify effectors.
Four of the effector proteins identified here (CBU1524, CBU1823, CBU1825 and CBU2052) were reported recently as Dot/Icm substrates in an independent study that was based on identifying effectors based on sequence motifs and protein interaction properties [41], however, the majority of the effectors identified using the type IV secretion signal screen were not uncovered using these predictions. This highlights the benefit of using functional screens for type IV secretion signals as a means to identify new effectors. It was also reported recently that the plasmid QpH1 in C. burnetii encodes several Dot/Icm effectors [42]. Interestingly, none of the effectors identified here using the type IV secretion signal screen were plasmid encoded. This may reflect a low abundance of QpH1 in the genomic DNA preparation used to make the Cya fusion protein library. Other reasons for effectors not being identified in this screen for type IV secretion signals include a lack of suitable SauIIIA sites that would be needed for in an in-frame fusion to Cya, and the potential to miss effectors translocated at low levels because the screening approach relied on a strong signal being generated by a single clone in a mixed pool. These limitations should be considered in any future screens for type IV secretion signals, and with slight modifications to the DNA fragmentation method and screening protocol, minor improvements should enable more complete coverage of the genome.
An analysis was performed in which the effectors found in the NM strain were compared to genes from other sequenced strains of C. burnetii. As was observed for members of the Ank family of effector proteins [25], a significant degree of heterogeneity was revealed among the Dot/Icm effectors identified in this study. Interestingly, what appears to be effector pseudogenization is prevalent in strains of C. burnetii capable of causing acute and chronic Q fever in humans. The evolutionary and pathogenic significance toward Dot/Icm effector pseudogenization remains unclear. Dugway possesses the largest genome and fewest pseudogenes of the sequenced C. burnetii isolates [31]. This implies that a large repertoire of functional effectors could be beneficial, as it may enable C. burnetii to persist and be transmitted by many different animals in nature without causing disease to their hosts. The loss of effectors appears to correlate with a change in the relationship between C. burnetii and mammalian hosts from commensalism to parasitism, which would indicate that a balanced repertoire of effectors is important in preventing responses that would result in host pathology.
Further investigation is required to determine whether the truncated NM Dot/Icm effectors identified here are non-functional or whether these genes still produce functional effectors that mediate the same or distinct activities compared to the larger proteins predicted to be produced by alleles found in other strains of C. burnetii. Because effectors are modular proteins that can have multiple enzymatic domains with different effector functions, it remains possible that some of the mutations resulting in truncation of a NM protein occur through positive selection, where the truncated effector has evolved in such a way that it retains a beneficial enzymatic activity and discards an activity that is no longer being selected for in the infected host. CBU0080 and the homologue CbuK1976 may provide such an example. Ectopically expressed CbuK1976 has an affinity for the host nucleus, whereas, the truncated NM protein CBU0080, which lacks a helix loop helix motif found in CbuK1976, is retained in the cytoplasm. Conceivably, CBU0080 could be a functional effector where elimination of the helix loop helix motif is the result of positive selection as it may enhance an activity associated with the C-terminal region of the protein. The effectors CBU1107 and CBU1108 provide another intriguing comparison. CBU1107 and CBU1108 have independent determinants that confer Dot/Icm-dependent translocation. In Dugway, genes encoding these two effectors are fused to generate the large protein CBUD1210, which would represent a hybrid effector. It is possible that a mutation in NM resulting in two gene products, CBU1107 and CBU1108, was positively selected because it allows more precise regulation of two effector functions compared to the hybrid protein CBUD1210. Alternatively, CBUD1210 could represent fusion of the two genes encoding CBU1107 and CBU1108 and this fusion was positively selected because it links two effectors that may act in concert. Although these examples remain highly speculative, it illustrates how studies on C. burnetii effector evolution could provide novel insight into how the repertoire of Dot/Icm effectors is being shaped by host interactions.
Several approaches were used to investigate whether the identified effector proteins had activities that would perturb eukaryotic cell functions (summarized in Table 1). C. burnetii Dot/Icm effector proteins are likely to modulate a broad array of eukaryotic cellular processes. This is supported by the diversity observed in the localization of these effectors produced in mammalian host cells. Although several effectors were found diffusely in the cytosol of the mammalian cell, there were clear examples of specific localization of effectors to the nucleus, mitochondria, lysosomes and a perinuclear region near the Golgi apparatus. Consistent with its localization near the Golgi apparatus, the effector CBU0635 disrupted membrane transport in the host secretory pathway as determined by a defect in SEAP secretion. The localization of CBU0077 with lysosomes was also intriguing, as the CCV is an organelle derived from lysosomes. The observation that CBU0077 is highly conserved in all sequenced strains of C. burnetii makes this protein an attractive candidate for being directly involved in processes important for CCV biogenesis. Isolation of a C. burnetii mutant deficient in CBU0077 may provide some clues as to the role this effector plays in modulating host processes during infection.
Six of the C. burnetii effectors were found to moderately affect the replication of yeast when overproduced. These differences were minor compared to what is observed when many of the L. pneumophila effectors are produced in S. cerevisiae, where replication and viability are affected so severely that the yeast are unable to form single colonies on agar plates. Because of the severity of these defects, several L. pneumophila Dot/Icm effector proteins were identified based on their capacity to interfere with yeast viability [37]. Furthermore, examination of the impact of a large cohort of L. pneumophila Dot/Icm effectors showed that in excess of 60% of these proteins significantly decrease yeast growth and/or viability [38]. The observation that none of the C. burnetii effectors had this dramatic effect on yeast viability could relate to the different roles the Dot/Icm system is likely to play during infection by these two pathogens. For L. pneumophila, the Dot/Icm system has evolved to modulate cellular functions in evolutionarily diverse protozoan hosts and to create a vacuole that is derived from the endoplasmic reticulum. Many of the targets that have been identified for L. pneumophila effectors are proteins that are highly conserved throughout the eukaryotic kingdom. By contrast, C. burnetii infects mammals and has reprogrammed its effector repertoire to survive and replicate inside mammalian host cells. As such, there are likely to be effectors that have evolved to manipulate targets specific to mammalian cells and may not recognize targets in more primitive eukaryotic cells, such as yeast. An example of this is the effector AnkG that blocks apoptosis by targeting the host protein p32, which is found in mammals but not in yeast or protozoa [26].
Studies on the effector CBU0077 showed that this protein was produced during infection. Both the endogenous CBU0077 protein and the BlaM-CBU0077 fusion protein were delivered into host cells. Because the BlaM detection system is based on delivery of a robust enzyme into the host cytosol, the assay is sensitive and can detect translocation of small amounts of a fusion protein. Detectible levels of effector translocation were first detected at 8 h post infection. The inability to detect translocation of BlaM-CBU0077 at 4 h suggests that the C. burnetii Dot/Icm system is not fully functional during the early stages of host cell infection. Bacterial proton motive force is involved in the translocation of effectors by the L. pneumophila Dot/Icm system [43], which implies that bacterial metabolism is important for the functioning of this apparatus. It has been shown that infectious forms of C. burnetii that are environmentally resistant do not become metabolically active until they gain access to a low pH environment [3], [44], [45]. Thus, it is possible that the Dot/Icm system is not functioning in the early events in vacuole biogenesis and only becomes active once the C. burnetii have been transported through the endocytic pathway of the host cell to an acidified lysosomal compartment. There are several logical reasons why the Dot/Icm system may not be needed by C. burnetii during these early stages of infection. Because fusion of phagosomes with lysosomes represents a default membrane transport pathway that most bacteria follow after uptake, the Dot/Icm system should not be needed to promote delivery of the bacteria to this preferred location. Additionally, it is now well established that mammalian cells have several innate immune sensors that can detect the activities of the L. pneumophila Dot/Icm system during the early stages of infection [46], [47]. Thus, not having the Dot/icm system functioning during these early infection events may help delay or prevent innate immune detection of C. burnetii.
Recent advances in axenic cultivation of C. burnetii [48] and newly developed genetic tools developed to study this organism [40] were used to isolate a transposon insertion mutant that lacked a functional Dot/Icm apparatus. As expected, the BlaM-CBU0077, BlaM-CBU0635 and BlaM-CBU1524 proteins were not delivered into host cells by the icmL::Tn mutant, providing additional evidence that translocation of these effectors by C. burnetii requires a functional Dot/Icm system. Similar to what was shown initially for L. pneumophila dot and icm mutants [12], [13], the ability of the C. burnetii icmL::Tn mutant to grow on synthetic medium as efficiently as the isogenic parental strain demonstrates conclusively that the Dot/Icm system is not essential for replication extracellularly. Most importantly, when the icmL::Tn was used to infect mammalian cells, there was a severe defect in the ability of this mutant to replicate intracellularly that was linked genetically to the inactivation of the Dot/Icm system by insertion of the transposon.
These data demonstrate that the Dot/Icm system plays an essential role in the process of intracellular infection of mammalian host cells, which means that the collective activities of the effector proteins delivered into cells by the Dot/Icm system are needed for the successful establishment and maintenance of the specialized vacuole in which this pathogen resides. Thus, determining the biochemical activities of the novel C. burnetii effectors identified in this study should help elucidate the mechanisms by which this intracellular pathogen can persist and grow inside mammalian host cells.
Methods
Reagents
Unless otherwise noted, chemicals were purchased from Sigma (St. Louis, MO). Restriction enzymes and molecular cloning enzymes were purchased from New England Biolabs (NEB; Ipswich, MA). Cell culture media and fetal bovine serum (FBS) was obtained from Invitrogen (Carlsbad, CA). Protease inhibitor cocktail and Fugene 6 transfection reagent were from Roche Applied Sciences (Indianapolis, IN).
Host cell lines and bacterial strains
CHO cells expressing FcγRII [28], HeLa 229, Vero and HEK 293 cells were maintained in Dulbecco's Modified Eagle's Media (DMEM) supplemented with 10% heat-inactivated FBS at 37°C in 5%CO2 unless otherwise described.
A plaque-purified isolate of Coxiella burnetii phase II Nine Mile strain, a generous gift from Dr. T. Hackstadt of the Rocky Mountain Laboratories (Hamilton, MT) was propagated in eukaryotic cell lines in DMEM supplemented with 5% FBS at 37°C in 5% CO2 or ACCM at 37°C in 5% CO2 and 2.5% O2 as described previously [48]. When required chloramphenicol and kanamycin were used in C. burnetii ACCM cultures at 3 µg/ml and 275 µg/ml respectively.
Legionella pneumophila strains, CR39 and CR58 ( ΔdotA), which are isogenic derivatives of the serogroup 1 strain LP01 [13], [49], were grown on either charcoal-yeast extract (CYE) plates or in ACES-buffered yeast extract (AYE) broth containing chloramphenicol (6.25 µg/ml) as previously described [50]. E. coli strains were cultivated on Luria-Bertani (LB) plates or broth supplemented with chloramphenicol (25 µg/ml), ampicillin (100 µg/ml) or kanamycin (50 µg/ml) as appropriate.
Construction of C. burnetii genomic DNA library
Purified C. burnetii were pelleted for 5 min at 8000×g in a microcentrifuge. Pellets were resuspended in 50 mM Tris-HCl, pH 8.0 containing 50 mM EDTA, pH 8.0. Lysozyme was added to a final concentration of 5 mg/ml and incubated at RT with gentle rocking. After 30 min, sodium dodecyl sulfate (SDS) and proteinase K (Roche Applied Sciences) were added to final concentrations of 1% and 10 mg/ml, respectively, and incubated for 1 h at RT with gentle rocking. Genomic DNA was extracted with phenol, phenol-chloroform, and chloroform; precipitated with ethanol; air dried; then resuspended in 10 mM Tris-HCl, pH 8.0 containing 1 mM EDTA, pH 8.0. Genomic DNA was partially digested with Sau3AI for 1 h at 37°C, separated by gel electrophoresis, and purified from a Low Melt agarose gel (American Bioanalytical, Natick, MA) as follows. DNA fragments between 2 kb and 4 kb were excised from the gel, placed in eppendorf tubes, and heated to 70°C for 10 min. An equal volume of prewarmed phenol (40°C) was added to the melted agarose, mixed, and separated by 5 min centrifugation at 16,000×g. The upper aqueous layer was chloroform extracted, ethanol precipitated and resuspended in 10 mM Tris-HCl, pH 8.0 containing 1 mM EDTA, pH 8.0.
Plasmid, pEC33, was generated from pM45-ralF [27] as follows: plasmid was digested with BamHI and PstI, gel purified to remove ralF gene insert, treated with DNA polymerase I (Klenow) fragment, and ligated to preserve the BamHI site (E. Cambronne, unpublished).
C. burnetii genomic DNA fragments were ligated into BamHI digested pEC33 resulting in a library of C. burnetii genes fused to the carboxyl region of the enzymatic domain of the Bordetella pertussis adenylate cyclase toxin. The genomic library was transformed into ElectroMax DH10B electroporation competent E. coli (Invitrogen). The insert efficiency was estimated to be 92% by restriction enzyme digest of randomly selected clones. Following amplification in E. coli, the library DNA was purified by cesium chloride density fractionation and subsequently used to transform L. pneumophila.
Screening of genomic DNA library
To screen the C. burnetii genomic DNA library for translocation into mammalian host cells using the Cya fusion strategy [51], L. pneumophila strain CR39 was transformed with the library of C. burnetii genes fused to cya by electroporation and the library of transformants was expanded on CYE plates. The library was distributed into pools containing approximately 50 transformants in each, and the pools were expanded by growing the bacteria on 24 well plates of CYE supplemented with chloramphenicol for 4 days at 37°C. Bacteria from each well were resuspended in AYE medium, an equal volume of freeze media (4% (w/v) peptone containing 10% (v/v) glycerol) was added, and the pools were stored in a 96-well plate format at −80°C.
Briefly, the translocation assay was performed as follows (see Figure S2). Pools of L. pneumophila expressing Cya-C. burnetii fusion proteins growing on CYE in 24 well plates were resuspended in AYE to a concentration of 1×109 cfu/ml. CHO - FcγRII cells were pretreated for 1 h with an anti-Legionella antibody then infected with 3×106 L. pneumophila from each pool. After 1 h of incubation at 37°C, monolayers were washed three times with ice-cold PBS and lysed in cold buffer containing 50 mM HCl and 0.1% (v/v) TX-100 for 30 min at 4°C. The lysates were boiled for 5 min and neutralized with 30 mM NaOH. Total cAMP was extracted and quantified using cAMP Biotrak Enzymeimmunoassay System (GE Healthcare, Piscataway, NJ). Pools that registered cAMP levels of 2.5 times over vector control were considered positive.
Individual colonies from pools that were scored positive in the cAMP assay were isolated and arrayed in 96 well plates containing CYE agar. Secondary screening was conducted by pooling the 12 clones in each row to identify which row contained a positive clone (Figure S2B). Rows that gave a positive cAMP signal were further analyzed by testing individual clones present in the positive rows for translocation (Figure S2C).
Plasmid DNA from individual L. pneumophila transformants that were identified using the above screening procedure was isolated as follows. Bacteria from a confluent region on a CYE plate were used to inoculate AYE containing chloramphenicol and grown overnight at 37°C. Bacteria were pelleted and resuspended in STET buffer (8% (w/v) sucrose, 50 mM Tris-HCl, pH 8.0, 50 mM EDTA, pH 8.0, 0.1% (v/v) Triton X-100). Fresh lysozyme (2 mg/ml) was added and incubated for 5 min at RT before boiling the samples for 1 min. Plasmid DNA was isolated from aqueous phase using Miniprep Express Matrix (Bio101, La Jolla, CA) as per manufacturer's instructions. To sequence fusion junctions, plasmid DNA was isolated from E. coli transformants as described above and sequenced by W.M Keck Foundation Biotechnology Resource Laboratory (Yale University).
Plasmid construction
Full-length genes of constructs containing in-frame fusions of putative effectors were amplified by PCR with sequence specific primers containing BamHI and PstI sites for cloning into pEC33 (Table S3). pcDNA4/TO (Invitrogen) with a N-terminal 3×FLAG tag (pFLAG) was used to clone Dot/Icm effectors from NM, Dugway, G and K isolates through BamHI and XhoI or BamHI and EcoRI for CBU0295 and CBU1823 or BamHI and XbaI for CBU2059 (Table S3). Plasmids encoding in frame fusions between the coding regions of BlaM and the effector proteins were generated by inserting the gene encoding the effector at a SalI site located at the 3′ end of the blaM gene in the plasmid pJB-CAT-BlaM [42] using the In-Fusion Advantage PCR Cloning system as described by the manufacturer (Clontech Laboratories, Palo Alto, CA). For complementation studies the icmL gene and the icmQPONL2L1KEGC operon were ligated into the plasmid pJB-CAT-BlaM after digestion of the DNA with PstI and SalI to remove the blaM gene, generating the plasmids pIcmL and pQC, respectively.
RT-PCR analysis
Total RNA was extracted from C. burnetii purified from persistently infected CHO - FcγRII cells using a Trizol Max Bacterial Isolation kit (Invitrogen) as per manufacturer's instructions. RNA was treated with RNase-free DNase (Qiagen, Valencia, CA) and purified using RNeasy mini spin columns (Qiagen) as per manufacturer's instructions for RNA cleanup. First strand cDNA was synthesized from 5 µg total RNA using gene specific RT primers (Table S3) with the Superscript III First Strand Synthesis System for RT-PCR (Invitrogen). PCR reactions were performed from cDNA prepared with or without Superscript III reverse transcriptase using gene specific primers and Elongase enzyme mix (Invitrogen).
Fluorescence microscopy
To examine the localization of ectopically produced FLAG-tagged proteins, HeLa 229 or CHO - FcγRII cells were plated on 12 mm glass coverslips in 24-well plates at a density of 3×104 cells/well. Cells were transfected with plasmid DNA using Fugene 6 (Roche) or Lipofectamine 2000 (Invitrogen). Eighteen hours post-transfection cells were fixed using 4% paraformaldehyde and permeablized with Triton X-100. Samples were incubated with primary antibodies in 2% BSA at the following concentrations; anti-FLAG (Sigma) 1∶300, anti-LAMP1 H4A3 (Developmental Studies Hybridoma Bank) 1∶50, anti-TOM22 (Sigma) 1∶250, anti-GM130 (BD) 1∶250, anti-C. burnetii 1∶10000. Secondary antibodies, Alexa Fluor 488 and 546 (Invitrogen) were used at 1∶2000 in 2% BSA. Bacterial and host cell DNA was labeled using 0.1 µg/ml 4,6-diamidino-2-phenylindole (DAPI). Coverslips were mounted on slides using ProLong Gold (Invitrogen). Images were acquired with a Zeiss LSM510 microscope using a 100×/1.4 numerical aperture objective. Images were exported as TIFF files and labeled with Adobe Illustrator. Similar results were obtained in at least three independent experiments.
CBU0077 translocation assay
Uninfected or C. burnetii-infected HeLa cells were incubated with chloramphenicol (25 µg/ml) or DMSO for 16 h at 37°C in 5%CO2. Cells were washed with Hanks buffered salt solution (HBSS, Invitrogen) containing 1× protease inhibitor cocktail (PI) then lysed with ice-cold HBSS containing PI and 0.1% Triton X-100. The cells were centrifuged for 10 min at 16,000g at 4°C to separate the soluble fraction containing secreted and translocated bacterial proteins from the insoluble fraction, which contained intact bacteria. The soluble fraction was clarified using a 0.22 µm pore-size filter (Millipore, Bedford, MA) and proteins were concentrated by trichloroacetic acid precipitation. Samples from both the soluble and insoluble fractions were separated by SDS-PAGE, transferred to PVDF (Millipore) and probed with anti-CBU0077 or anti-EF-Ts [52] as a bacterial lysis control. Similar results were obtained from three independent experiments.
Yeast growth assay
Genes encoding Dot/Icm effectors were amplified from NM gDNA by PCR with sequence specific primers containing a BamHI site, Kozak sequence and ATG, if applicable, at the 5′ end, and an XbaI or SphI at the 3′end (Table S3). The PCR products were cloned into pYES2 (Invitrogen), and introduced in competent S.cerevisiae. Overnight liquid YNB-glucose cultures were pelleted and washed once with YNB before being resuspended to an OD 600 nm of approximately 0.03 in YNB-galactose. 200 µl of this suspension was distributed into duplicate wells of a 96 well tray and incubated in a Tecan M1000 plate reader at 30°C. OD 600 nm readings were taken every 30 minutes, following a 30 second agitation, for the duration of the time course. Growth curves were created from at least three independent experiments.
SEAP assay
HEK 293 cells in 24 well plates were co-transfected with 0.2 µg pSEAP and 0.3 µg of a pFLAG derivative encoding the indicated Dot/Icm effector or control protein. Approximately 16 h post-transfection the cells were washed with fresh medium, and a sample was collected to calculate the background SEAP activity in the culture supernatant. Following an 8 h incubation period, the culture medium was collected to assay SEAP levels secreted by the cells, and an equivalent volume of buffer containing 0.2% Triton X-100 was applied to the cells to measure the intracellular alkaline phosphatase activity. Alkaline phosphatase activity was measured in 96-well format using the Phospha-Light System (Applied Biosystems). Ratios of extracellular to intracellular SEAP activity were calculated from at least three independent experiments.
Genetic manipulation of C. burnetii
Plasmid DNA, for introduction into C. burnetii, was purified using the GeneElute HP Endotoxin Free Plasmid Maxiprep Kit (Sigma) and concentrated with an Amicon Ultra Centrifugal Filter (Millipore). This DNA was introduced into axenically grown C. burnetii by electroporation. Approximately 5×109 bacteria were washed twice and resuspended in ice-cold 10% glycerol. Plasmid DNA was added (10 µg) and the bacteria were electroporated at 18 kV, 500Ω, 25 µF. Electroporated C. burnetii were resuspended in 1 ml RPMI and 200 µl of this was used to seed a 20 ml ACCM culture. After a 24 h recovery, the appropriate antibiotics were added. Cultures were passaged after 7 days. Following 14 days of selection in liquid ACCM the bacteria were grown on ACCM agarose plates to isolate individual transformants.
Southern hybridization
PCR probes for icmL and the transposon were amplified, with the incorporation of digoxigenin-dUTP, using the primer pairs icmL.1F/icmL.1R and TnF/TnR respectively (Table S3). gDNA was digested with HindIII, separated by agarose gel electrophoresis and transferred to positively charged nylon membrane. Hybridization was performed overnight at 60°C and DIG detection was performed according to the manufacturers' guidelines (Roche).
BlaM translocation assay
CBU0077, CBU0635 and CBU1524 were introduced into the SalI site of pJB-CAT-BlaM to create a BlaM fusion constructs under the control of the CBU1169 promoter. These constructs were electroporated into C. burnetii and individual clones isolated after two passages with chloramphenicol selection. BlaM expression was confirmed by western blot with anti-BlaM (1∶1000, QED Bioscience Inc, San Diego, CA). Approximately 2×104 HeLa cells were seeded in black clear bottom 96 well trays (Corning Incorporated, Corning NY). The next day monolayers were infected with C. burnetii at the appropriate MOI and the infection allowed to proceed until the desired time point. Cells were loaded with the fluorescent substrate CCF4/AM, using the LiveBLAzer-FRET B/G Loading Kit (Invitrogen) with 15 mM probenecid, in the dark for 2 h at room temperature. Fluorescence, using an excitation of 415 nm and emission at 460 nm and 535 nm, was quantified using the Tecan M1000 plate reader. Following background subtraction the ratio of 460 nm to 535 nm (blue∶green) was calculated and expressed relative to uninfected cells. For single cell assays, the infection and stain procedure was similar. Cells were visualized by fluorescence microscopy using an inverted Nikon Eclipse TE-2000 S microscope and a 20× objective. A total of 400 cells were counted in three independent wells to determine the percent of cells that were BlaM positive. Images were acquired, exported as TIFF files and labeled with Adobe Illustrator.
C. burnetii infections
HeLa 229 and Vero cells were plated at a density of 5×104 into 24 well trays in DMEM with 2%FBS. Axenically grown C. burnetii strains were quantified by qPCR using dotA specific primers [3] to provide an accurate MOI of 50. Following a 4 h infection period, cells were washed once and incubated with fresh DMEM with 2% FBS. Wells were collected for analysis 24 (Day 1), 72 (Day 3), 120 (Day 5) and 168 (Day 7) h after this initial time point. Cells were either fixed with 4% paraformaldehyde for subsequent immunofluorescent analysis or lysed by hypotonic lysis and collected for C. burnetii quantification. Bacteria were pelleted from lysed cells, gDNA extracted using the illustra bacteria genomicPrep Mini Prep Kit (GE Healthcare) and genomic equivalents quantified by qPCR. To quantify the proportion of cells intracellular after the 4 h infection period fixed cells were stained with mouse anti-C. burnetii (1∶5000) and Alexa Fluor 596 anti-mouse before secondary fixation and permeablization. Samples were then stained with rabbit anti-C. burnetii (1∶10000) and Alexa Fluor 488 anti-rabbit. The proportion of cell associated bacteria that were intracellular, those stained green only, were quantified for at least 400 HeLa cells per coverslip. Two coverslips per sample were quantified from three independent experiments.
For the intracellular viability studies the infections with C. burnetii were performed as described above. At the indicated time points after infection the HeLa cells were extensively washed with PBS to remove any remaining extracellular bacteria before hypotonic lysis. The lysed cell material was added to 1 ml of ACCM and then diluted 1∶100 in ACCM. Genome equivalents in the starting samples were determined by qPCR. The diluted lysates in ACCM were incubated at 37°C, 5% CO2, 2.5% O2 for seven days and qPCR was used to determine the amount of replication that had occurred.
Supporting Information
Zdroje
1. MaurinMRaoultD 1999 Q fever. Clin Microbiol Rev 12 518 553
2. HackstadtTWilliamsJC 1981 Biochemical stratagem for obligate parasitism of eukaryotic cells by Coxiella burnetii. Proc Natl Acad Sci U S A 78 3240 3244
3. ColemanSAFischerERHoweDMeadDJHeinzenRA 2004 Temporal analysis of Coxiella burnetii morphological differentiation. J Bacteriol 186 7344 7352
4. HoweDMallaviaLP 2000 Coxiella burnetii exhibits morphological change and delays phagolysosomal fusion after internalization by J774A.1 cells. Infect Immun 68 3815 3821
5. HoweDHeinzenRA 2006 Coxiella burnetii inhabits a cholesterol-rich vacuole and influences cellular cholesterol metabolism. Cell Microbiol 8 496 507
6. RomanoPSGutierrezMGBeronWRabinovitchMColomboMI 2007 The autophagic pathway is actively modulated by phase II Coxiella burnetii to efficiently replicate in the host cell. Cell Microbiol 9 891 909
7. HoweDMelnicakovaJBarakIHeinzenRA 2003 Maturation of the Coxiella burnetii parasitophorous vacuole requires bacterial protein synthesis but not replication. Cell Microbiol 5 469 480
8. SeshadriRPaulsenITEisenJAReadTDNelsonKE 2003 Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc Natl Acad Sci U S A 100 5455 5460
9. ZamboniDSMcGrathSRabinovitchMRoyCR 2003 Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system. Mol Microbiol 49 965 976
10. ZusmanTYerushalmiGSegalG 2003 Functional similarities between the icm/dot pathogenesis systems of Coxiella burnetii and Legionella pneumophila. Infect Immun 71 3714 3723
11. SegalGShumanHA 1999 Legionella pneumophila utilizes the same genes to multiply within Acanthamoeba castellanii and human macrophages. Infect Immun 67 2117 2124
12. MarraABlanderSJHorwitzMAShumanHA 1992 Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc Natl Acad Sci U S A 89 9607 9611
13. BergerKHIsbergRR 1993 Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol 7 7 19
14. BrandBCSadoskyABShumanHA 1994 The Legionella pneumophila icm locus: a set of genes required for intracellular multiplication in human macrophages. Mol Microbiol 14 797 808
15. RoyCRBergerKHIsbergRR 1998 Legionella pneumophila DotA protein is required for early phagosome trafficking decisions that occur within minutes of bacterial uptake. Mol Microbiol 28 663 674
16. KaganJCRoyCR 2002 Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat Cell Biol 4 945 954
17. KaganJCSteinMPPypaertMRoyCR 2004 Legionella subvert the functions of Rab1 and Sec22b to create a replicative organelle. J Exp Med 199 1201 1211
18. BursteinDZusmanTDegtyarEVinerRSegalG 2009 Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog 5 e1000508
19. HuangLBoydDAmyotWMHempsteadADLuoZQ 2010 The E Block motif is associated with Legionella pneumophila translocated substrates. Cell Microbiol 13 227 245
20. MurataTDelpratoAIngmundsonAToomreDKLambrightDG 2006 The Legionella pneumophila effector protein DrrA is a Rab1 guanine nucleotide-exchange factor. Nat Cell Biol 8 971 977
21. MachnerMPIsbergRR 2006 Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev Cell 11 47 56
22. MullerMPPetersHBlumerJBlankenfeldtWGoodyRS 2010 The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science 329 946 949
23. NagaiHKaganJCZhuXKahnRARoyCR 2002 A bacterial guanine nucleotide exchange factor activates ARF on Legionella phagosomes. Science 295 679 682
24. PanXLuhrmannASatohALaskowski-ArceMARoyCR 2008 Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science 320 1651 1654
25. VothDEHoweDBearePAVogelJPUnsworthN 2009 The Coxiella burnetii ankyrin repeat domain-containing protein family is heterogeneous, with C-terminal truncations that influence Dot/Icm-mediated secretion. J Bacteriol 191 4232 4242
26. LuhrmannANogueiraCVCareyKLRoyCR 2010 Inhibition of pathogen-induced apoptosis by a Coxiella burnetii type IV effector protein. Proc Natl Acad Sci U S A 107 18997 19001
27. NagaiHCambronneEDKaganJCAmorJCKahnRA 2005 A C-terminal translocation signal required for Dot/Icm-dependent delivery of the Legionella RalF protein to host cells. Proc Natl Acad Sci U S A 102 826 831
28. JoinerKAFuhrmanSAMiettinenHMKasperLHMellmanI 1990 Toxoplasma gondii: fusion competence of parasitophorous vacuoles in Fc receptor-transfected fibroblasts. Science 249 641 646
29. LuoZQIsbergRR 2004 Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci U S A 101 841 846
30. StoennerHGLackmanDB 1960 The biologic properties of Coxiella burnetii isolated from rodents collected in Utah. Am J Hyg 71 45 51
31. BearePAUnsworthNAndohMVothDEOmslandA 2009 Comparative genomics reveal extensive transposon-mediated genomic plasticity and diversity among potential effector proteins within the genus Coxiella. Infect Immun 77 642 656
32. CazaletCRusniokCBruggemannHZidaneNMagnierA 2004 Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet 36 1165 1173
33. SchultzJMilpetzFBorkPPontingCP 1998 SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci U S A 95 5857 5864
34. CirilloSLLumJCirilloJD 2000 Identification of novel loci involved in entry by Legionella pneumophila. Microbiology 146 1345 1359
35. LiuMConoverGMIsbergRR 2008 Legionella pneumophila EnhC is required for efficient replication in tumour necrosis factor alpha-stimulated macrophages. Cell Microbiol 10 1906 1923
36. NewtonHJSansomFMDaoJMcAlisterADSloanJ 2007 Sel1 repeat protein LpnE is a Legionella pneumophila virulence determinant that influences vacuolar trafficking. Infect Immun 75 5575 5585
37. CampodonicoEMChesnelLRoyCR 2005 A yeast genetic system for the identification and characterization of substrate proteins transferred into host cells by the Legionella pneumophila Dot/Icm system. Mol Microbiol 56 918 933
38. HeidtmanMChenEJMoyMYIsbergRR 2009 Large-scale identification of Legionella pneumophila Dot/Icm substrates that modulate host cell vesicle trafficking pathways. Cell Microbiol 11 230 248
39. de FelipeKSGloverRTCharpentierXAndersonORReyesM 2008 Legionella eukaryotic-like type IV substrates interfere with organelle trafficking. PLoS Pathog 4 e1000117
40. BearePAHoweDCockrellDCOmslandAHansenB 2009 Characterization of a Coxiella burnetii ftsZ mutant generated by Himar1 transposon mutagenesis. J Bacteriol 191 1369 1381
41. ChenCBangaSMertensKWeberMMGorbaslievaI 2010 Large-scale identification and translocation of type IV secretion substrates by Coxiella burnetii. Proc Natl Acad Sci U S A 107 21755 60
42. VothDEBearePAHoweDSharmaUMSamoilisG 2011 The Coxiella burnetii Cryptic Plasmid is Enriched in Genes Encoding Type IV Secretion System Substrates. J Bacteriol 193 1493 1503
43. CharpentierXGabayJEReyesMZhuJWWeissA 2009 Chemical genetics reveals bacterial and host cell functions critical for type IV effector translocation by Legionella pneumophila. PLoS Pathog 5 e1000501
44. HackstadtTWilliamsJC 1983 pH dependence of the Coxiella burnetii glutamate transport system. J Bacteriol 154 598 603
45. OmslandACockrellDCFischerERHeinzenRA 2008 Sustained axenic metabolic activity by the obligate intracellular bacterium Coxiella burnetii. J Bacteriol 190 3203 3212
46. ShinSCaseCLArcherKANogueiraCVKobayashiKS 2008 Type IV secretion-dependent activation of host MAP kinases induces an increased proinflammatory cytokine response to Legionella pneumophila. PLoS Pathog 4 e1000220
47. ArcherKARoyCR 2006 MyD88-dependent responses involving toll-like receptor 2 are important for protection and clearance of Legionella pneumophila in a mouse model of Legionnaires' disease. Infect Immun 74 3325 3333
48. OmslandACockrellDCHoweDFischerERVirtanevaK 2009 Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc Natl Acad Sci U S A 106 4430 4434
49. ZuckmanDMHungJBRoyCR 1999 Pore-forming activity is not sufficient for Legionella pneumophila phagosome trafficking and intracellular growth. Mol Microbiol 32 990 1001
50. FeeleyJCGibsonRJGormanGWLangfordNCRasheedJK 1979 Charcoal-yeast extract agar: primary isolation medium for Legionella pneumophila. J Clin Microbiol 10 437 441
51. SoryMPCornelisGR 1994 Translocation of a hybrid YopE-adenylate cyclase from Yersinia enterocolitica into HeLa cells. Mol Microbiol 14 583 594
52. SeshadriRHendrixLRSamuelJE 1999 Differential expression of translational elements by life cycle variants of Coxiella burnetii. Infect Immun 67 6026 6033
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článek Distribution of the Phenotypic Effects of Random Homologous Recombination between Two Virus SpeciesČlánek SIV Nef Proteins Recruit the AP-2 Complex to Antagonize Tetherin and Facilitate Virion ReleaseČlánek Dual Function of the NK Cell Receptor 2B4 (CD244) in the Regulation of HCV-Specific CD8+ T CellsČlánek A Large and Intact Viral Particle Penetrates the Endoplasmic Reticulum Membrane to Reach the CytosolČlánek Interleukin-13 Promotes Susceptibility to Chlamydial Infection of the Respiratory and Genital Tracts
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2011 Číslo 5- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Lymphoadenopathy during Lyme Borreliosis Is Caused by Spirochete Migration-Induced Specific B Cell Activation
- Infections Are Virulent and Inhibit the Human Malaria Parasite in
- A Gamma Interferon Independent Mechanism of CD4 T Cell Mediated Control of Infection
- MDA5 and TLR3 Initiate Pro-Inflammatory Signaling Pathways Leading to Rhinovirus-Induced Airways Inflammation and Hyperresponsiveness
- The OXI1 Kinase Pathway Mediates -Induced Growth Promotion in Arabidopsis
- An E2F1-Mediated DNA Damage Response Contributes to the Replication of Human Cytomegalovirus
- Quantitative Subcellular Proteome and Secretome Profiling of Influenza A Virus-Infected Human Primary Macrophages
- Distribution of the Phenotypic Effects of Random Homologous Recombination between Two Virus Species
- Inhibition of Both HIV-1 Reverse Transcription and Gene Expression by a Cyclic Peptide that Binds the Tat-Transactivating Response Element (TAR) RNA
- A Viral Satellite RNA Induces Yellow Symptoms on Tobacco by Targeting a Gene Involved in Chlorophyll Biosynthesis using the RNA Silencing Machinery
- Misregulation of Underlies the Developmental Abnormalities Caused by Three Distinct Viral Silencing Suppressors in Arabidopsis
- Investigating the Host Binding Signature on the PfEMP1 Protein Family
- Human Neutrophil Clearance of Bacterial Pathogens Triggers Anti-Microbial γδ T Cell Responses in Early Infection
- Septation of Infectious Hyphae Is Critical for Appressoria Formation and Virulence in the Smut Fungus
- A Family of Helminth Molecules that Modulate Innate Cell Responses via Molecular Mimicry of Host Antimicrobial Peptides
- Phospholipids Trigger Capsular Enlargement during Interactions with Amoebae and Macrophages
- CTL Escape Mediated by Proteasomal Destruction of an HIV-1 Cryptic Epitope
- Evolution of Th2 Immunity: A Rapid Repair Response to Tissue Destructive Pathogens
- Extensive Genome-Wide Variability of Human Cytomegalovirus in Congenitally Infected Infants
- The Antiviral Efficacy of HIV-Specific CD8 T-Cells to a Conserved Epitope Is Heavily Dependent on the Infecting HIV-1 Isolate
- Epstein-Barr Virus Infection of Polarized Epithelial Cells via the Basolateral Surface by Memory B Cell-Mediated Transfer Infection
- Reactive Oxygen Species Hydrogen Peroxide Mediates Kaposi's Sarcoma-Associated Herpesvirus Reactivation from Latency
- Crystal Structure and Functional Analysis of the SARS-Coronavirus RNA Cap 2′-O-Methyltransferase nsp10/nsp16 Complex
- The Dot/Icm System Delivers a Unique Repertoire of Type IV Effectors into Host Cells and Is Required for Intracellular Replication
- AAV Exploits Subcellular Stress Associated with Inflammation, Endoplasmic Reticulum Expansion, and Misfolded Proteins in Models of Cystic Fibrosis
- Suboptimal Activation of Antigen-Specific CD4 Effector Cells Enables Persistence of In Vivo
- SIV Nef Proteins Recruit the AP-2 Complex to Antagonize Tetherin and Facilitate Virion Release
- Dual Function of the NK Cell Receptor 2B4 (CD244) in the Regulation of HCV-Specific CD8+ T Cells
- Transition of Sporozoites into Liver Stage-Like Forms Is Regulated by the RNA Binding Protein Pumilio
- A Large and Intact Viral Particle Penetrates the Endoplasmic Reticulum Membrane to Reach the Cytosol
- Transcriptome Analysis of in Human Whole Blood and Mutagenesis Studies Identify Virulence Factors Involved in Blood Survival
- Interleukin-13 Promotes Susceptibility to Chlamydial Infection of the Respiratory and Genital Tracts
- Structural Insights into Viral Determinants of Nematode Mediated Transmission
- Protective Efficacy of Serially Up-Ranked Subdominant CD8 T Cell Epitopes against Virus Challenges
- Viral CTL Escape Mutants Are Generated in Lymph Nodes and Subsequently Become Fixed in Plasma and Rectal Mucosa during Acute SIV Infection of Macaques
- Taking Some of the Mystery out of Host∶Virus Interactions
- Viral Small Interfering RNAs Target Host Genes to Mediate Disease Symptoms in Plants
- : An Emerging Cause of Sexually Transmitted Disease in Women
- Mitochondrial Ubiquitin Ligase MARCH5 Promotes TLR7 Signaling by Attenuating TANK Action
- The Hexamer Structure of the Rift Valley Fever Virus Nucleoprotein Suggests a Mechanism for its Assembly into Ribonucleoprotein Complexes
- Acquisition of Human-Type Receptor Binding Specificity by New H5N1 Influenza Virus Sublineages during Their Emergence in Birds in Egypt
- Stromal Down-Regulation of Macrophage CD4/CCR5 Expression and NF-κB Activation Mediates HIV-1 Non-Permissiveness in Intestinal Macrophages
- A Component of the Xanthomonadaceae Type IV Secretion System Combines a VirB7 Motif with a N0 Domain Found in Outer Membrane Transport Proteins
- Perturbs Iron Trafficking in the Epithelium to Grow on the Cell Surface
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Crystal Structure and Functional Analysis of the SARS-Coronavirus RNA Cap 2′-O-Methyltransferase nsp10/nsp16 Complex
- Lymphoadenopathy during Lyme Borreliosis Is Caused by Spirochete Migration-Induced Specific B Cell Activation
- The OXI1 Kinase Pathway Mediates -Induced Growth Promotion in Arabidopsis
- : An Emerging Cause of Sexually Transmitted Disease in Women
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



