-
Články
- Časopisy
- Kurzy
- Témy
- Kongresy
- Videa
- Podcasty
An Apicoplast Localized Ubiquitylation System Is Required for the Import of Nuclear-encoded Plastid Proteins
Apicomplexan parasites are responsible for numerous important human diseases including toxoplasmosis, cryptosporidiosis, and most importantly malaria. There is a constant need for new antimalarials, and one of most keenly pursued drug targets is an ancient algal endosymbiont, the apicoplast. The apicoplast is essential for parasite survival, and several aspects of its metabolism and maintenance have been validated as targets of anti-parasitic drug treatment. Most apicoplast proteins are nuclear encoded and have to be imported into the organelle. Recently, a protein translocon typically required for endoplasmic reticulum associated protein degradation (ERAD) has been proposed to act in apicoplast protein import. Here, we show ubiquitylation to be a conserved and essential component of this process. We identify apicoplast localized ubiquitin activating, conjugating and ligating enzymes in Toxoplasma gondii and Plasmodium falciparum and observe biochemical activity by in vitro reconstitution. Using conditional gene ablation and complementation analysis we link this activity to apicoplast protein import and parasite survival. Our studies suggest ubiquitylation to be a mechanistic requirement of apicoplast protein import independent to the proteasomal degradation pathway.
Published in the journal: An Apicoplast Localized Ubiquitylation System Is Required for the Import of Nuclear-encoded Plastid Proteins. PLoS Pathog 9(6): e32767. doi:10.1371/journal.ppat.1003426
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1003426Summary
Apicomplexan parasites are responsible for numerous important human diseases including toxoplasmosis, cryptosporidiosis, and most importantly malaria. There is a constant need for new antimalarials, and one of most keenly pursued drug targets is an ancient algal endosymbiont, the apicoplast. The apicoplast is essential for parasite survival, and several aspects of its metabolism and maintenance have been validated as targets of anti-parasitic drug treatment. Most apicoplast proteins are nuclear encoded and have to be imported into the organelle. Recently, a protein translocon typically required for endoplasmic reticulum associated protein degradation (ERAD) has been proposed to act in apicoplast protein import. Here, we show ubiquitylation to be a conserved and essential component of this process. We identify apicoplast localized ubiquitin activating, conjugating and ligating enzymes in Toxoplasma gondii and Plasmodium falciparum and observe biochemical activity by in vitro reconstitution. Using conditional gene ablation and complementation analysis we link this activity to apicoplast protein import and parasite survival. Our studies suggest ubiquitylation to be a mechanistic requirement of apicoplast protein import independent to the proteasomal degradation pathway.
Introduction
Apicomplexans are eukaryotic pathogens and responsible for important human and animal diseases including malaria and toxoplasmosis. The Apicomplexa evolved from single-celled photosynthetic algae, and their adaptation to animal parasitism likely predates the emergence of animals from water to land. The presence of a plastid, the apicoplast, is the most important remnant of this evolutionary past [1], [2]. While no longer photosynthetic, the organelle synthesizes isoprenoids and fatty acids [3]. The apicoplast is essential for parasite survival, and its metabolism, biogenesis and maintenance are important targets for anti-parasitic drug treatment. The apicoplast was derived by secondary endosymbiosis, where a unicellular red alga was incorporated into a heterotrophic protist. As a consequence of this secondary endosymbiosis the apicoplast is surrounded by four membranes. The organelle carries a genome, yet most of its proteins are nuclear-encoded and imported into the organelle after translation. Targeting depends on a bipartite leader peptide, the first section of which mediates co-translational import into the endoplasmic reticulum, and the second part mediates delivery to the apicoplast, likely through fusion of endosomal vesicles with the outermost membrane of the organelle [4]. Three translocons breaching successive membranes have been proposed to act in the further transport of proteins into the stroma of the apicoplast [5]. The two inner membranes of the apicoplast are homologous to the membranes of the primary chloroplast and protein transport depends on systems derived from the chloroplast TIC and TOC machinery [6], [7], [8], [9]. Insight into the third translocon emerged first in cryptomonads, an algal group that like Apicomplexa harbors a secondary plastid. The secondary plastids of cryptomonads retained a remnant of the algal nucleus, the nucleomorph. Analysis of the gene content of the nucleomorph led to the discovery of plastid proteins that resembled components of the endoplasmic reticulum associated degradation (ERAD) machinery [10]. ERAD is a quality control mechanism that retro-translocates misfolded secretory proteins across the ER membrane [11]. Sommer and colleagues proposed that this mechanism has been adapted for protein import in secondary plastids [10]. There is now significant support for this hypothesis. Homologs of ERAD proteins have been identified and localized to plastids in various algal and apicomplexan species including a core of the membrane protein Der1, the AAA ATPase Cdc48 and its cofactor Ufd1 [10], [12], [13], [14], [15]. Recombinant plastid proteins can complement yeast ERAD mutants [14]. Importantly, genetic ablation of the ERAD component Der1Ap in T. gondii blocks apicoplast protein import, producing a phenotype that closely resembles ablation of the apicoplast TIC component Tic20 [6], [15].
During classical ERAD, protein translocation coincides with ubiquitylation, a process that typically employs a cascade of three enzymes: ubiquitin-activating enzyme (E1), ubiquitin-conjugating enzyme (E2), and ubiquitin ligase (E3) [16], [17]. Consuming ATP, the E1 enzyme adenylates ubiquitin at the C-terminus, creating a mixed anhydride. The sulfhydryl group of the E1 active-site cysteine then attacks the anhydride, which results in the formation of a high-energy thio-ester linking ubiquitin to E1. Ubiquitin is then passed to the active-site cysteine of the E2 enzyme. Lastly, with the aid of an E3 ligase, ubiquitin is transferred from E2 and covalently attached to the ε-amino group of a lysine in the target protein. Although clearly important in mediating ERAD, the role of ubiquitylation in protein import into secondary plastids is unclear. Interestingly, some ERAD-like ubiquitylation factors are observed in the plastids of cryptomonads, diatoms, and Apicomplexa [12], [18], [19].
While protein degradation is the key function of classical ERAD this could seem counterintuitive in the context of apicoplast protein import. However ubiquitin's functions are not limited to proteasomal degradation and extend to a variety of cellular protein trafficking systems [20]. Furthermore, ubiquitylation may be a critical mechanistic requirement of protein transport via the ERAD translocon [11], [21]. Some authors now view the ERAD associated E3 ligase Hrd1 as a favored candidate for the actual protein-conducting pore [22].
In this study, we elucidate the function of ubiquitylation in the apicoplast. We identify and localize a comprehensive set of ubiquitylating components in the apicomplexan parasites P. falciparum and T. gondii. Using recombinant apicoplast enzymes from P. falciparum we reconstitute ubiquitylation in vitro using a variety of heterologous and homologous cofactors. By genetic analysis in T. gondii we demonstrate that loss of the apicoplast-localized ubiquitin-conjugating enzyme leads to loss of apicoplast protein import and parasite demise. Importantly complementation of this mutant depends on an active site cysteine required for enzymatic activity. Taken together our experiments suggest an essential mechanistic role for the ERAD-like ubiquitylation machinery in apicoplast protein import.
Results
Ubiquitylation factors localize to the apicoplast in Toxoplasma and Plasmodium
Using a combination of computational approaches we identified a comprehensive set of proteins that may act as apicoplast ubiquitylation system (see Materials and Methods). The results of these analyses (summarized in Table S1 in Text S1) identified apicoplast candidates for E1, E2 and E3 enzymes in both P. falciparum and T. gondii. We next determined whether these candidates are indeed targeted to the apicoplast. We targeted the locus of T. gondii TgE1Ap by single homologous integration and placed a haemagglutinin (HA) epitope tag at the C-terminus of the protein. Stable transgenic clones show apicoplast staining when labeled with an anti-HA antibody by immunofluorescence (Fig. 1A, the P. falciparum homolog E1 is also localized to the apicoplast [12]). Our attempts to localize the candidates for apicoplast E2 by tagging the respective genes directly in the locus did not produce viable transgenics in either T. gondii or P. falciparum. Epitope fusion close to the C-terminal active domain may interfere with function and prevent replacement of the native gene. However, the coding sequence of TgE2Ap could be fused to an epitope tag in the context of an ectopic expression plasmid (maintaining the native locus). Parasites expressing this construct show apicoplast labeling indistinguishable from that observed for E1 when probed with an epitope specific antibody. To localize the Plasmodium homolog (and to aid subsequent biochemical analysis) we also expressed a portion of Mal13P1.227 fused to an affinity tag in E. coli and used the purified recombinant protein to raise a specific antiserum. Immunofluorescence assays on P. falciparum parasites with this serum produced labeling that coincides with labeling for the apicoplast marker ACP (Fig. 1 C).
Fig. 1. Ubiquitylation proteins localize to the apicoplast in Plasmodium falciparum and Toxoplasma gondii. 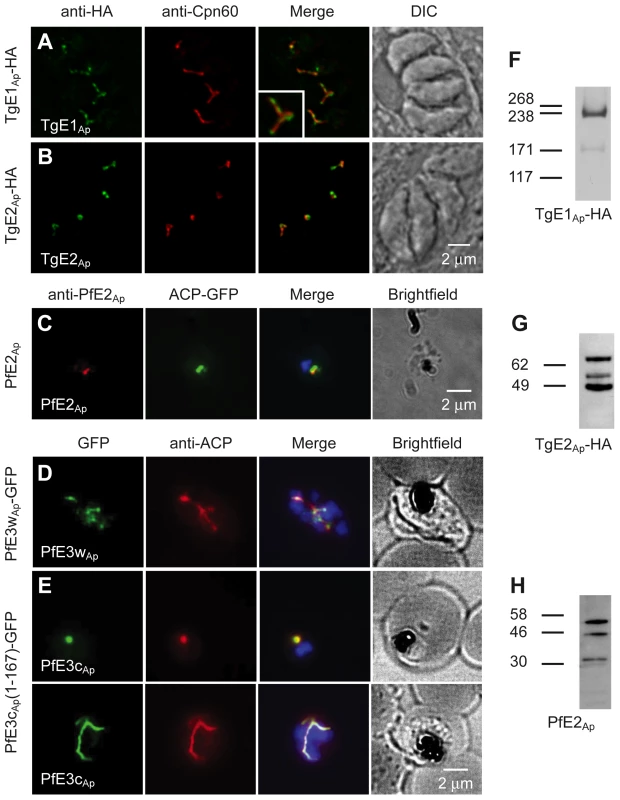
(A–E) Immunofluorescence assays detecting the respective ubiquitylation factor indicated (white lettering) in the left-most panel. Proteins were detected by tagging with an HA or GFP epitope in the genomic locus (A, D), by ectopic fusion with full (B) or partial coding sequence (E), or by using an antibody raised again recombinant protein (C). Staining for the apicoplast markers Cpn60 and ACP is shown in the second lane. Merged images also show DAPI staining for P. falciparum. Insert in (B) shows a 200% enlargement. Western blots of T. gondii (F, G, transgene as indicated) or P. falciparum (H) protein samples reacted with anti-HA and anti-PfE2Ap antibodies. Predicted sizes of initial translation product of TgE2Ap, PfE2Ap and TgE1Ap are 70.4 kDa, 32.8 kDa and 315.9 kDa respectively. Two putative apicoplast E3 ubiquitin ligases were identified in Plasmodium, PfE3cAp (PFC0740c - PF3D7_0316900) and PfE3wAp (PFC0510w - PF3D7_0312100), and two in Toxoplasma (TGME49_226740 and TGME49_304460). We attempted to tag the proteins by placing different epitopes at the C-terminus through homologous gene targeting but were not successful. In case of PfE3cAp transgenics that showed initial locus targeting were quickly lost upon selection (Fig. S1A in Text S1). However, we recovered viable transgenic parasites tagged in the PfE3wAp locus. Targeted integration of the cassette and transcription of PfE3wAp-GFP was confirmed by PCR and RT-PCR (Fig. S1B–C in Text S1). Immunofluorescence assays showed PfE3wAp-GFP to localize to the apicoplast (Fig. 1D). Finally, using an episomal expression vector, we found that the first 167 amino acids of PfE3cAp target a GFP reporter to the apicoplast (Fig. 1E).
Apicoplast proteins are often processed at the N-terminus removing a leader peptide [4]. We analyzed processing for TgE1Ap, TgE2Ap and PfE2Ap for which suitable reagents were available. TgE1Ap produces the pattern typical for apicoplast proteins, two major bands likely corresponding to the precursor (heavier band) and mature protein (lighter band) Fig. 1F. Interestingly both TgE2Ap and PfE2Ap blots showed additional bands potentially arising from further post-translational modification (Fig. 1 G, H).
While the immunofluorescence assays indicate apicoplast localization of the ubiquitylation enzymes, overlap with luminal markers is only partial (see enlarged insert in Fig. 1A). We fixed and processed TgE2AP-HA parasites for electron microscopy and incubated cryo-sections with an anti-HA antibody. Note that gold particles are found in the membranous periphery of the apicoplast (Fig. 2 and Fig. S4 in Text S1). This labeling is indistinguishable from that previously observed for the apicoplast ERAD-like proteins Der1Ap and Cdc48Ap [15] and the periplastid protein PPP1 [23]. We conclude that the apicoplast has a full complement of E1, E2 and E3 ubiquitylation enzymes localized to the periphery of the organelle, most likely the periplastid compartment as observed for the ERAD-like system in the diatom Phaeodactylum tricornitum [14], [18], [19].
Fig. 2. TgE2Ap is localized to the periphery of the apicoplast. 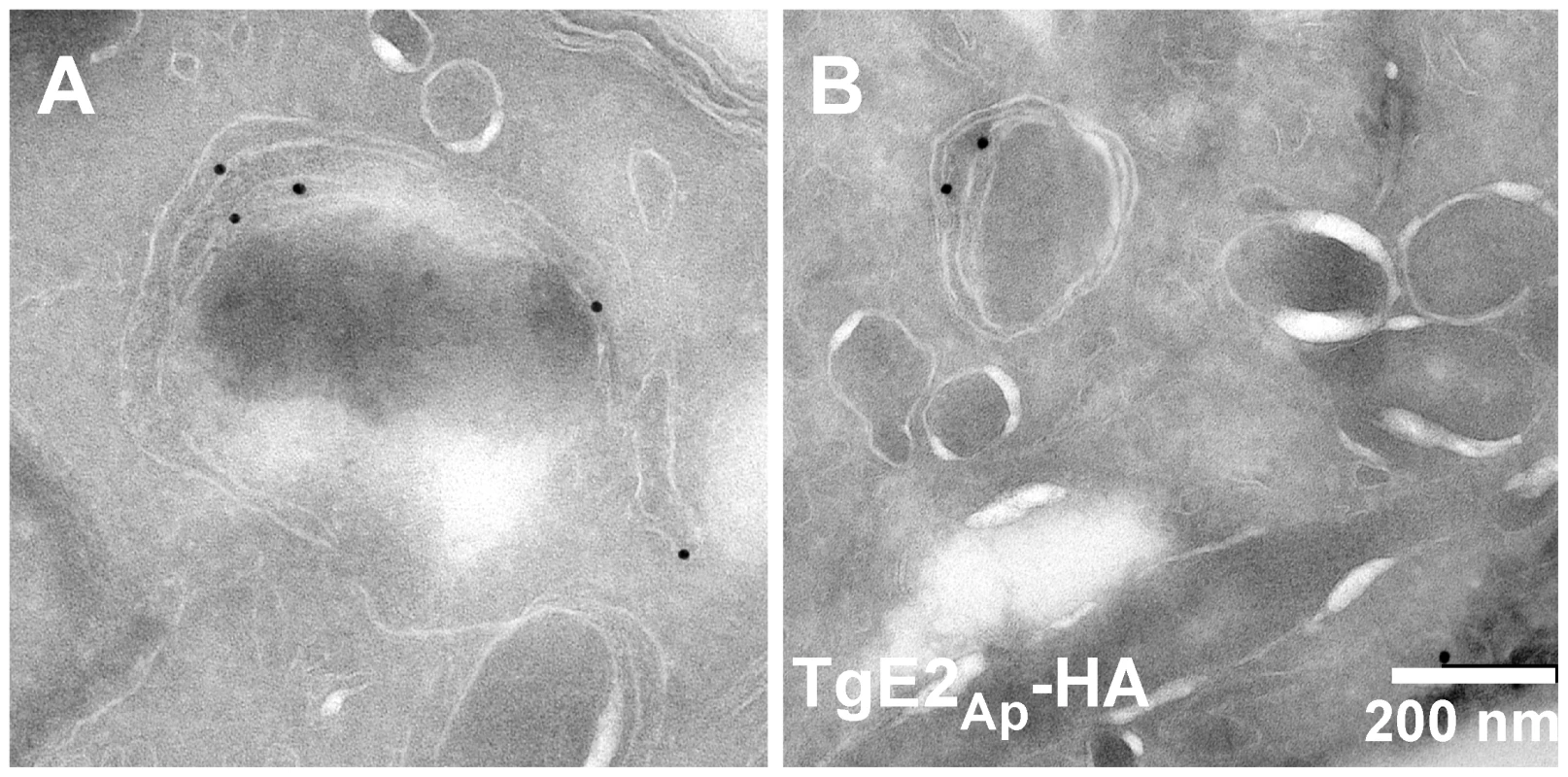
(A, B) Transmission electron micrograph of cryo-sections prepared from the TgE2Ap -HA parasite line and labeled with anti-HA antibody and antiglobulin conjugated to colloidal gold. Gold beads are observed at the periphery of an organelle that is surrounded by four membranes. In vitro reconstitution of ubiquitylation using recombinant and native Plasmodium apicoplast enzymes
We next sought to establish whether the candidate apicoplast ubiquitylation system is capable of activating and ligating ubiquitin. We amplified or synthesized sequences encoding full length PfE1LAp and PfE2Ap, or the RING domains of PfE3wAp and PfE3cAp respectively, and engineered them to be expressed as recombinant fusion proteins carrying an N-terminal glutathione S-transferase (GST) and/or six-histidine (HIS) affinity tag. Proteins of the expected size could be purified for all four constructs (Fig. 3A, B). We established biochemical ubiquitylation assays using combinations of parasite enzymes and commercially available heterologous components (Fig. 3C, recombinant human factors are shown in red, Plasmodium enzymes in green). Enzymes were incubated with recombinant human ubiquitin in a buffer containing an ATP regenerating system. When analyzed by Western blot, ubiquitin chains can be detected as ladders of high molecular weight bands [24]. Among the numerous human ubiquitin-activating enzymes tested, UBCH5a and UBCH13 were found to be suitable partners for PfE3cAp and PfE3wAp leading to robust ubiquitylation. Note that this activity is strictly dependent on the recombinant parasite E3 and absent in controls (Fig. 3D, E). The pattern obtained differed between the two E2 enzymes and suggested ubiquitylation of the RING domain in the context of only UBCH5a, while interaction with UBCH13 appeared to produce free poly-ubiquitin. Variation of ubiquitylation pattern depending on the E2 partnered with the ligase is frequently observed [25]. To test this independently we probed the in vitro reaction with anti-GST antibody to visualize the E3 and its higher molecular weight ubiquitin adducts. Consistently, this revealed shifts in molecular weight of PfE3cAp and PfE3wAp only when incubated with UBCH5a (Fig. 3F) as free polyubiquitin is not detected in this assay format.
Fig. 3. Recombinant Plasmodium falciparum apicoplast proteins show ubiquitin activation, conjugation and ligation activity. 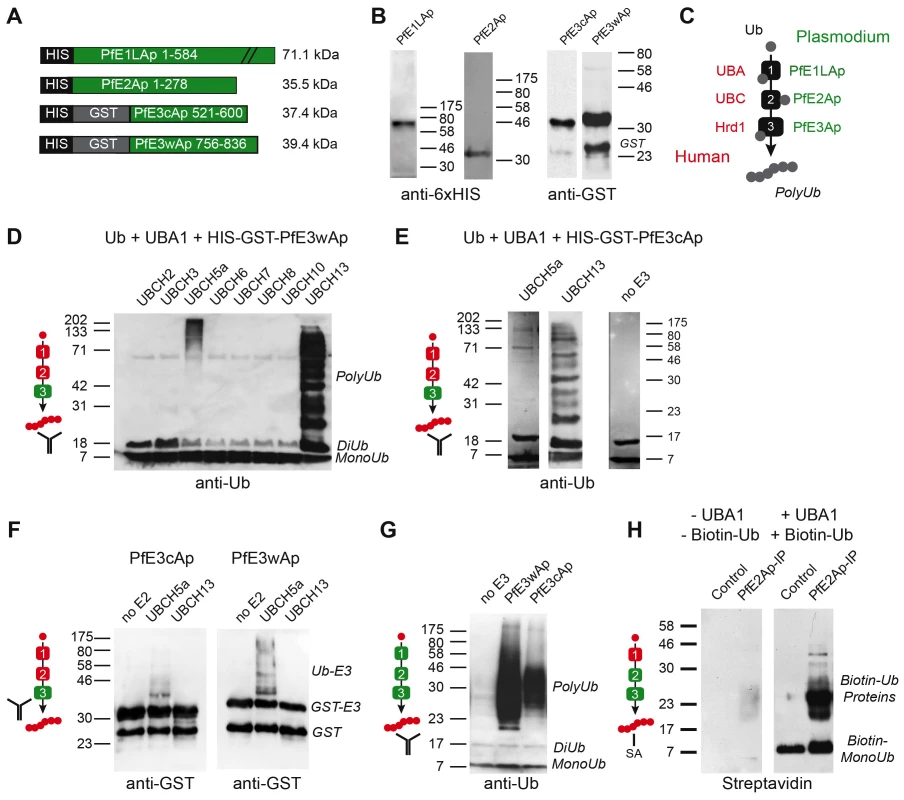
(A) Schematic maps of recombinant proteins indicating protein domains and purification tags used and the predicted molecular weight of the fusion protein (not to scale). (B) Western blot of recombinant proteins detected with antibodies to the indicated tag. (C) Schematic outline of the biochemical assay, ubiquitin is incubated with recombinant E1, E2 and E3 enzymes in the presence of ATP, human enzymes are shown in red, parasite enzymes in green (this color scheme is used as reference in panels D–H). Ubiquitylation is measured by Western blot detected polyubiquitin chains (PolyUb) either free, or linked to the E3 RING domain. The antibody symbol indicates the specific protein detected in each panel. PfE3wAp (D) or PfE3cAp (E) were incubated with human UBA1 and different UBCs as indicated. Note ubiquitylation using UBC5a (E3 associated higher molecular weight) and UBC13 (free Ub chains, only selected panels are shown in E). (F) Only the use of UBC5a results in ubiquitylation attached to PFE3Ap visible as higher molecular weight species recognized by the antibody to the GST tag of E3. (G) Reconstitution of ubiquitylation using recombinant Plasmodium E1Ap and E2Ap and PfE3wAp or PfE3cAp respectively. (H) The presumptive E2/E3 complex immunoprecipitated from P. falciparum parasites using an antibody to PfE2Ap shows ubiquitylation activity when incubated with biotinylated human ubiquitin and UBA1 (detected with Streptavidin, SA). Next we tested whether ubiquitylation activity can be reconstituted entirely with parasite enzymes. When recombinant PfE1LAp and PfE2Ap were incubated with ubiquitin alone (Fig. 3G, left lane), no ubiquitylation was detected. However, upon addition of recombinant E3 ligase PfE3wAp or PfE3cAp, ubiquitylation was readily observed. Lastly we wished to evaluate the activity for native parasite enzymes. Among the reagents generated and tested in this study a custom-made antibody to PfE2Ap was found suitable for immunoprecipitation under native conditions. Often the conjugating and ligating enzymes form a complex and can be co-precipitated and detected by their combined activity [26], [27]. We incubated pull down fractions from parasite lysates with recombinant human UBA1, and biotinylated-ubiquitin (using tagged ubiquitin enhances sensitivity and focuses the assay on only newly ubiquitylated proteins). We observed significant ubiquitylation that was dependent on the immunoprecipitate and UBA1 (Fig. 3H). Taken together our observations provide biochemical support for the notion that the apicoplast ERAD-like system is capable of mediating ubiquitylation.
E2Ap is required for parasite growth and protein import into the apicoplast
The apicoplast ERAD system has a critical role in protein import into the organelle [5], [18]. We tested whether ubiquitylation is a mechanistic requirement of this process by genetic ablation of the apicoplast ERAD-like ubiquitylation enzymes. We attempted disruption of the loci of PfE3cAp, PfE3wAp, and PfsUBA1. We isolated strains bearing drug marker insertions in the PfE3wAp gene and documented loss of associated transcription (Fig. S2B, S3C in Text S1). However, we also noted multiple genomic duplications in these strains complicating interpretation (Fig. S3D in Text S1). We did not obtain viable parasites with disrupted PfE3cAP or PfsUBA1 loci. This is consistent with a potentially essential role for these proteins, and we therefore turned to T. gondii where the construction of conditional mutants is feasible.
We engineered a parasite strain where the endogenous promoter of the TgE2Ap gene was replaced by a regulatable promoter in the following referred to as (i)ΔTgE2Ap (Fig. 4A, [23]). This was accomplished by double cross over in the T. gondii TATiΔKu80 background, a parasite line that favors homologous recombination and expresses a transactivator that can be modulated using anhydrotetracycline (ATc). Drug resistant parasite clones were tested by PCR and integration of the promoter was confirmed by Southern blot. We monitored the level of TgE2Ap mRNA in response to ATc by quantitative PCR. Fig. 4D shows down-regulation of the transcript below 10% of its normal level at day four of ATc treatment. We asked whether loss of TgE2Ap affects parasite growth and performed plaque and real-time fluorescence assays. Parasites grow normally in the absence of ATc indicated by formation of plaques, however in the presence of ATc, plaque formation is severely attenuated (Fig. 4F). Similarly, (i)ΔTgE2Ap parasites show significant growth reduction in the fluorescence assay in the presence of ATc (Fig. 4E), preincubation of parasites in ATc abolished growth entirely. We conclude, that TgE2Ap is critical for parasite growth.
Fig. 4. Genetic ablation of E2Ap in T. gondii results in a block of apicoplast protein import and parasite growth. 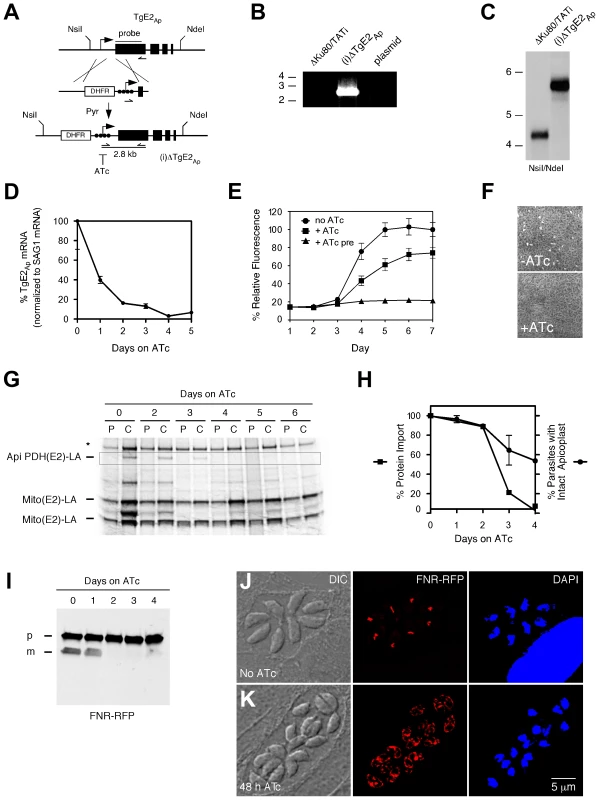
(A) The TgE2Ap locus was modified homologous recombination and insertion of a DHFR selectable marker and a regulatable promoter (arrow, black circles indicate tet operator elements). Positions of diagnostic primers (half arrows) and probe (black line) are indicated. (B) PCR test for insertion, note that only the recombined locus is suitable to template the 2.8 kbp product. (C) Southern analysis hybridizing genomic DNA form parental (ΔKu80/TATI) or mutant ((i)ΔTgE2Ap) parasites with a probe derived from the first exon of TgE2Ap. (D) Real time PCR analysis of TgE2Ap expression in (i)ΔTgE2Ap parasites upon ATc treatment (normalized to the mRNA of the major surface protein SAG1, untreated control set to 100%, error bars show SD, n = 3) detects the expected DNA fragments. PCR analysis of clones shows integration of the promoter. (E) Growth of (i)ΔTgE2Ap was measured by fluorescence (circles no drug, squares ATc, triangles 3 days of pretreatment with ATc prior to plate inoculation) and (F) plaque assay. (G) Apicoplast protein import in (i)ΔTgE2Ap was measured by following PDH(E2) lipoylation ( [6] highlighted by box). Note loss of band upon ATc treatment and persistence of mitochondrial lipoylation (Mito (E2)-LA); Please note that there are two lipoylated proteins in the T. gondii mitochondrion [42], [43], * human PDH-E2. (H) Quantification of protein import (as measured in G, squares, data shown is representative of four experiments) and number of apicoplasts per parasite (circles, n = 3, SD shown) in (i)ΔTgE2Ap over the course of ATc treatment. (I) Western blots probing FNR-RFP in (i)ΔTgE2Ap upon ATc treatment (p, precursor; m, mature protein). Note reduction in mature band upon treatment. Immunofluorescence assays showing (i)ΔTgE2Ap in the absence (J) or presence (K) of ATc. 38% of parasite vacuoles showed labeling outside the apicoplast, likely due to back up of cargo into the ER at 48 h of treatment (<3% in untreated parasites). We next tested the ability of (i)ΔTgE2Ap parasites to import apicoplast proteins in the absence or presence of ATc and measured the import-dependent lipoylation of the apicoplast pyruvate dehydrogenase E2 subunit [6]. (i)ΔTgE2Ap parasites were treated with ATc for different periods and pulse-labeled for one hour with [35S] methionine/cysteine. For the chase samples the radioactive isotope was removed, and cells were incubated for two additional hours in normal media. The samples were then used for immunoprecipitation with an anti-lipoic acid antibody followed by separation on SDS-PAGE. Treatment of cells with ATc for 2 days resulted in attenuation of import, leading to complete loss after 4 days (Fig. 4G, H). Lipoylation of two mitochondrial enzymes remained unaffected. We also monitored apicoplast loss, a frequent consequence of interference with apicoplast protein import [6], [15]. We observed a drop over time, but note that loss of import significantly precedes plastid loss. Loss of apicoplast protein import has also been shown to result in loss of leader peptide removal and backing up of precursor protein into the ER and other elements of the secretory pathway [6], [15], [28] We therefore measured the levels of precursor and processed mature form of the apicoplast reporter protein FNR-RFP [29].. We grew parasites for 0 to 4 days on ATc and performed Western blots using parasite protein extracts from each day. Probing these blots with an antibody against RFP revealed that precursor levels of FNR-RFP remained unchanged throughout the 4 days, while the mature protein was no longer detected after 2 days on ATc further supporting a strong import defect (Fig. 4I). We also monitored the localization of FNR-RFP in treated and untreated (i)ΔTgE2Ap parasites by immunofluorescence assay. In untreated parasites FRN-RFP is restricted to the apicoplast (Fig. 4K). After 48 hours of ATc treatment 38% of parasite vacuoles also show significant labeling outside of the apicoplast surrounding the nucleus likely representing the ER (Fig. 4J, untreated TgE2Ap or ATc treated wild type parasites showed such labeling in <3% of counted four cell vacuoles, n = 200). We conclude that apicoplast protein import is impaired in the absence of TgE2Ap.
A conserved cysteine residue in the active site of TgE2Ap is required for its function
Apicoplast ubiquitylation enzymes are capable of synthesizing ubiquitin chains in vitro, but is this activity required in vivo? To test this we established a complementation assay. The coding sequence of the TgE2Ap gene driven by a constitutive promoter was introduced into the uracil-phosphoribosyltransferase (UPRT) locus of the (i)ΔTgE2Ap mutant (Fig. 5C). Parasites were selected for the loss of UPRT activity using 5-fluorodeoxyuridine [30] and a clonal cell line that now constitutively expressed a second copy of TgE2Ap in the conditional knock down background was isolated. We confirmed correct integration by PCR (Fig. 5D). We tested the ability of this strain to form plaques when expression from the native locus is ablated by ATc treatment, and found that genetic complementation fully rescued growth (Fig. 5E).
Fig. 5. Active site residues are required for functional complementation of the TgE2Ap null mutant. 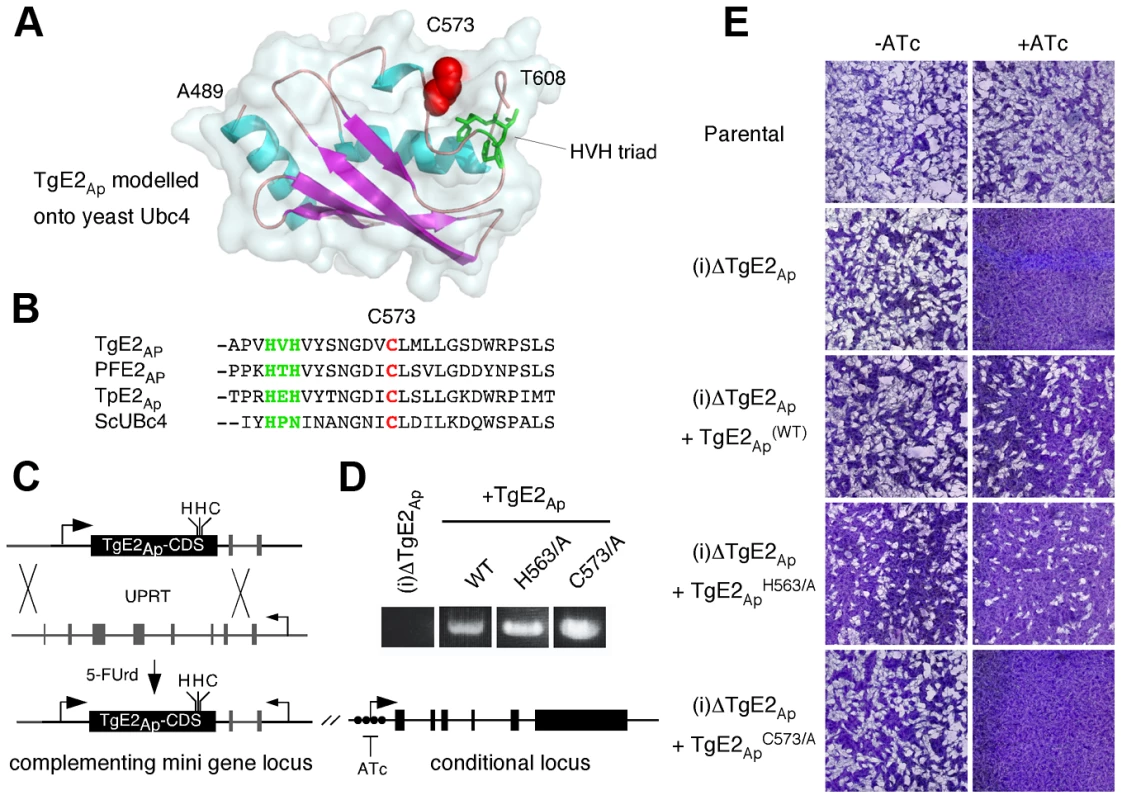
(A) MacPyMOL model of the secondary structure of TgE2Ap (alignable residues A489-T608) using S. cerevesiae Ubc-4 enzyme as template and. Note three conserved α-helices (blue) and four β-strands (magenta). The active site cysteine residue is shown in red the HVH triad in green. (B) Multiple sequence alignment of the active site (cysteine, red: triad, green) from P. falciparum, T. gondii, the diatom Thalassiosira pseudonana and yeast (complete alignment shown in Fig. S3 in Text S1). (C) Wild type and point mutants of the TgE2Ap coding sequence were introduced into the UPRT locus of TgE2Ap (C) under 5FUrd selection, correct insertion was established by PCR (D) and phenotypic complementation was assessed by plaque assay in the presence or absence of ATc (E). Multiple sequence alignment of TgE2Ap and E2 enzymes from a wide range of eukaryotes showed that TgE2Ap shares conserved features, reported earlier to be critical for this class of enzymes. We therefore modelled the C-terminal domain of TgE2Ap onto the structure of UBC4, a well characterize yeast ubiquitin conjugating enzyme [31]. Multiple sequence alignment and homology modelling identified C573 as the presumptive active site cysteine (Fig. 5A, B, see Fig. S3 in Text S1). Most E2 enzymes possess a signature HPN triad proximal to the active site cysteine [32]. The histidine has been previously suggested to be dispensable for E2-catalyzed ubiquitylation, but is important for the folding of the active site in other systems [33]. The asparagine residue on the other hand was consistently found to be important for RING-E3/E2-dependent ubiquitin conjugation [34]. A conserved HXH triad is found at this position in apicomplexans (Fig. 5B). We engineered a series of point mutants in TgE2Ap replacing C573, H563, and H565 with alanine respectively. These genes were then introduced into the (i)ΔTgE2Ap mutant as described above and tested for their ability to complement loss of TgE2Ap upon ATc treatment using plaque assay. Expression of the H563A point mutant fully complemented loss of native TgE2Ap (Fig. 5E) and parasites now grow even in the presence of ATc. In contrast, despite numerous attempts we were unable to establish a stable parasite line expressing H565A, which may suggest dominant effects of this mutation. We were able to isolate mutants expressing C573A, however these strains show no complementation, and are still fully susceptible to ATc treatment (Fig. 5E). We conclude that enzymatic activity is a requirement for TgE2Ap function in vivo and that C573 and H565 residues are critical for the function of the enzyme while H563 is likely dispensable.
Discussion
Endosymbiosis is a key evolutionary mechanism underlying the emergence and diversification of eukaryotes – in particular for photosynthetic eukaryotes. The acquisition of a eukaryotic red algal symbiont led to the chromalveolates, a large super-phylum of tremendous ecological diversity that includes apicomplexan parasites. The descendent of the algal symbiont, the apicoplast, maintains a highly compartmentalized organization, and nuclear encoded proteins have to overcome four membranes on their journey to the stroma. An apicoplast-localized ERAD-like system appears to play an important role in apicoplast protein import. Recent reports have identified and characterized components of this ERAD–like system in different algal and parasite species [7], [10], [12], [13], [14], [15]. In this study we provide significant biochemical and genetic evidence for the hypothesis that an apicoplast localized ubiquitylation cascade is an essential element of this protein import system. We identify apicoplast ubiquitin activating, conjugating and ligating enzymes in two important apicomplexan parasites, P. falciparum and T. gondii. We show in vitro and in vivo that these proteins have conserved biochemical activities and are capable of ubiquitin transfer. Finally, in genetic studies, we show that TgE2Ap, for which we were able to isolate a conditional mutant, is essential for apicoplast protein import, organellar maintenance and parasite growth. Overall these observations support a direct mechanistic role of ubiquitylation in protein translocation independent of ubiquitin's function in proteasomal degradation [11]. The classical ERAD system is believed to recognize and respond to the folding state of secretory proteins. Interestingly, recent studies show that the transit peptide of apicoplast proteins is primarily unstructured and that this conformation may be critical for proper transport to the organelle [35]. This model would need a distinguishing element to avoid elimination of apicoplast proteins by the classical ERAD. Specific chaperone sets could potentially provide such specificity, but remain to be discovered. A recent study in Arabidopsis has identified a role for ubiquitylation also in primary plastids, however this role appears to be distinct from secondary plastids. In this case ubiquitylation results in degradation of the components of the TOC complex and is thought to more globally regulate chloroplast biogenesis during plant development [36].
The identity of the apicoplast ubiquitin or ubiquitin-like modifier remains a significant unresolved question. Our results demonstrate that apicoplast enzymes are capable of acting on archetypical ubiquitin (recombinant human protein), studies in P. tricornitum show similar activity for a E3 ligase found in the diatom secondary chloroplast [18]. However whether the apicoplast system actually utilizes ubiquitin in vivo remains to be established. As shown in Fig. 1G and H Western blots for TgE2Ap and PfE2Ap show additional bands. It is conceivable that these bands represent ubiquitin or a ubiquitin-like protein covalently bound to the active site of the apicoplast localized E2. However we note that, for TgE2A, none of the bands was affected by reduction of the protein or point mutation of the active site cysteine (data not shown). Alternatively this may indicate an ubiquitin-like protein bound to a residue different from the active site of the enzyme or multiple processing steps as have been observed for some apicoplast membrane proteins [37]. Our efforts to demonstrate ubiquitin bound to apicoplast ubiquitylation enzymes purified from P. falciparum or T. gondii so far did not result in robust detection (using either antibodies or mass spectrometry, data not shown). Furthermore ubiquitylation of plastid-bound cargo proteins is not readily observed in apicomplexans or diatoms. A reasonable candidate for which apicoplast localization has been suggested [12] is an atypical, large ubiquitin-like protein (PF08_0067). Curiously, this protein lacks the di-glycine motif typically required for the formation of the isopeptide bond and a homolog has yet to be identified in the Toxoplasma genome. Similarly, plastid ubiquitin candidates from algae show lack of sequence motifs typically required for polyubiquitylation [19]. It is conceivable that this ubiquitin-like protein could be processed and/or ligated in a novel fashion that does not depend on a di-glycine sequence. Alternatively, its function may resemble that of the HERP protein in the classical ERAD pathway. Like PF08_0067, HERP has an ubiquitin-like domain at the N-terminus followed by transmembrane domains at the C-terminus [38]. HERP is believed to interact with HRD1 and to regulate the ubiquitylation activity of the ERAD translocon in response to folding stress [39]. In that case PF08_0067 is likely not the main substrate for the apicoplast ubiquitylation system and the modifier is yet to be discovered.
Studying the apicoplast ubiquitin faces technical obstacles that so far prevented direct tagging of the candidate ubiquitin and subsequent detection of modified cargo. There are several strong candidates for plastid-localized deubiquitylation enzyme in apicomplexans and diatoms (Table S1 in Text S1, [13], [18]). The activity of these enzymes may dramatically shorten the time ubiquitin remains on proteins and thus prevent the robust detection of ubiquitin adducts [22]. Isolation of mutants lacking apicoplast deubiquitylation might allow testing of this hypothesis and potentially lead to accumulation of modified cargo proteins. While a number of mechanistic details of the apicoplast ubiquitylation system remain to be elucidated, we demonstrate that the system is essential to the organelle and the parasite. Building on a longstanding effort to target ubiquitylation for the development of anti-cancer drugs [40] may potentially lead to new anti-parasitic compounds in the future.
Materials and Methods
P. falciparum strains 3D7, D10_ACP-(leader)-GFP (MR4, MRA568) and derivatives were cultured in human O+ red blood cells [41]. T. gondii RH strain parasites and derivatives were propagated in human fibroblasts and genetically modified as described [6], [15].
For in vitro ubiquitylation assays recombinant P. falciparum enzymes were incubated with recombinant human or parasite factors. Typically 50–200 µM recombinant ubiquitin, 0.05–0.2 µM E1 enzyme, 1–5 µM E2 enzymes, and 1–12.5 µM of E3 ligases were incubated in 50 mM Tris-HCl, pH 7.4, 1 mM DTT in presence of a re-energizing system (BostonBiochem) containing the ATP and ATP regenerating enzymes to recycle hydrolyzed ATP needed for the assay, for 2 hours at 37°C followed by SDS-PAGE and immunoblotting.
Ex vivo ubiquitylation assays were performed by lysing 3D7 P. falciparum in 20 mM HEPES pH 7.9, 10 mM KCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 0.5 mM AEBSF (Fisher Scientific), 0.65% Igepal v/v, and protease inhibitor cocktail (Roche), or 20 mM HEPES pH 7.9, 0.1 M NaCl, 0.1 mM EDTA, 0.1 mM EGTA, 1.5 mM MgCl2, 1 mM DTT, 1 mM AEBSF and protease inhibitor cocktail (Roche). Supernatants were pooled and proteins were precipitated using the indicated antibodies and magnetic Protein A beads. Proteins bound to beads were mixed with re-energizing buffer, 0.5 µg/µl biotin-conjugated ubiquitin, 5 mM AEBSF and protease inhibitor cocktail. Reactions were incubated at 30°C with gentle agitation for two hours. Samples were eluted with 4× Laemmli buffer and analysed using biotin affinity blots. Human recombinant UBE1 and UBC enzymes, E3 ligases biotin conjugated ubiquitin and re-energizing buffer used in these assays were purchased from Boston Biochem.
T. gondii gene models were tested by 5′ - and 3′-RACE. Note that additional exons were identified for TgE2Ap (see genbank JX431938 for correct sequence). A conditional TgE2Ap knock-out was generated by exchanging the native promoter for the tetracycline inducible t7s4 promoter in the TATiΔKu80 parasite background. The targeting construct used 1.2 kb up - and 1.5 kb downstream of the TgE2Ap start codon introduced into vector pDT7S4. Linearized plasmid was transfected into the parental strain followed by pyrimethamine selection. To complement the knock-out, a TgE2Ap minigene was inserted into the UPRT locus under the control of a constitutive sag1 promoter. Transgenics were isolated in 5 µM 5-FUDR and identified by PCR. Parasite growth was measured by fluorescence and plaque assay in the presence and absence of 0.5 µm anhydrotetracycline (ATc). Please refer to the supplement materials for a more detailed description of materials and methods used in this study (including a table of all primers).
Supporting Information
Zdroje
1. KohlerS, DelwicheCF, DennyPW, TilneyLG, WebsterP, et al. (1997) A plastid of probable green algal origin in Apicomplexan parasites. Science 275 : 1485–1489.
2. GouldSB, WallerRF, McFaddenGI (2008) Plastid evolution. Annual review of plant biology 59 : 491–517.
3. SeeberF, Soldati-FavreD (2010) Metabolic pathways in the apicoplast of apicomplexa. International review of cell and molecular biology 281 : 161–228.
4. WallerRF, KeelingPJ, DonaldRG, StriepenB, HandmanE, et al. (1998) Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proceedings of the National Academy of Sciences of the United States of America 95 : 12352–12357.
5. AgrawalS, StriepenB (2010) More membranes, more proteins: complex protein import mechanisms into secondary plastids. Protist 161 : 672–687.
6. van DoorenGG, TomovaC, AgrawalS, HumbelBM, StriepenB (2008) Toxoplasma gondii Tic20 is essential for apicoplast protein import. Proceedings of the National Academy of Sciences of the United States of America 105 : 13574–13579.
7. KalanonM, TonkinCJ, McFaddenGI (2009) Characterization of two putative protein translocation components in the apicoplast of Plasmodium falciparum. Eukaryotic cell 8 : 1146–1154.
8. BullmannL, HaarmannR, MirusO, BredemeierR, HempelF, et al. (2010) Filling the gap, evolutionarily conserved Omp85 in plastids of chromalveolates. The Journal of biological chemistry 285 : 6848–6856.
9. GlaserS, van DoorenGG, AgrawalS, BrooksCF, McFaddenGI, et al. (2012) Tic22 is an essential chaperone required for protein import into the apicoplast. The Journal of biological chemistry 287 : 39505–39512.
10. SommerMS, GouldSB, LehmannP, GruberA, PrzyborskiJM, et al. (2007) Der1-mediated preprotein import into the periplastid compartment of chromalveolates? Molecular biology and evolution 24 : 918–928.
11. SmithMH, PloeghHL, WeissmanJS (2011) Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science 334 : 1086–1090.
12. SporkS, HissJA, MandelK, SommerM, KooijTW, et al. (2009) An unusual ERAD-like complex is targeted to the apicoplast of Plasmodium falciparum. Eukaryotic cell 8 : 1134–1145.
13. PontsN, YangJ, ChungDW, PrudhommeJ, GirkeT, et al. (2008) Deciphering the ubiquitin-mediated pathway in apicomplexan parasites: a potential strategy to interfere with parasite virulence. PloS one 3: e2386.
14. HempelF, BullmannL, LauJ, ZaunerS, MaierUG (2009) ERAD-derived preprotein transport across the second outermost plastid membrane of diatoms. Molecular biology and evolution 26 : 1781–1790.
15. AgrawalS, van DoorenGG, BeattyWL, StriepenB (2009) Genetic evidence that an endosymbiont-derived endoplasmic reticulum-associated protein degradation (ERAD) system functions in import of apicoplast proteins. The Journal of biological chemistry 284 : 33683–33691.
16. PickartCM, FushmanD (2004) Polyubiquitin chains: polymeric protein signals. Current opinion in chemical biology 8 : 610–616.
17. LaneyJD, HochstrasserM (1999) Substrate targeting in the ubiquitin system. Cell 97 : 427–430.
18. HempelF, FelsnerG, MaierUG (2010) New mechanistic insights into pre-protein transport across the second outermost plastid membrane of diatoms. Molecular microbiology 76 : 793–801.
19. StorkS, MoogD, PrzyborskiJM, WilhelmiI, ZaunerS, et al. (2012) Distribution of the SELMA translocon in secondary plastids of red algal origin and predicted uncoupling of ubiquitin-dependent translocation from degradation. Eukaryotic cell 11 : 1472–1481.
20. MacgurnJA, HsuPC, EmrSD (2012) Ubiquitin and membrane protein turnover: from cradle to grave. Annual review of biochemistry 81 : 231–259.
21. YeY, MeyerHH, RapoportTA (2003) Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. The Journal of cell biology 162 : 71–84.
22. ErnstR, ClaessenJH, MuellerB, SanyalS, SpoonerE, et al. (2011) Enzymatic blockade of the ubiquitin-proteasome pathway. PLoS biology 8: e1000605.
23. SheinerL, DemerlyJL, PoulsenN, BeattyWL, LucasO, et al. (2011) A systematic screen to discover and analyze apicoplast proteins identifies a conserved and essential protein import factor. PLoS pathogens 7: e1002392.
24. ChengMC, HsiehEJ, ChenJH, ChenHY, LinTP (2012) Arabidopsis RGLG2, functioning as a RING E3 ligase, interacts with AtERF53 and negatively regulates the plant drought stress response. Plant physiology 158 : 363–375.
25. PetroskiMD, ZhouX, DongG, Daniel-IssakaniS, PayanDG, et al. (2007) Substrate modification with lysine 63-linked ubiquitin chains through the UBC13-UEV1A ubiquitin-conjugating enzyme. The Journal of biological chemistry 282 : 29936–29945.
26. VesterlundM, ZadjaliF, PerssonT, NielsenML, KesslerBM, et al. (2011) The SOCS2 ubiquitin ligase complex regulates growth hormone receptor levels. PloS one 6: e25358.
27. MerckxA, Le RochK, NivezMP, DorinD, AlanoP, et al. (2003) Identification and initial characterization of three novel cyclin-related proteins of the human malaria parasite Plasmodium falciparum. The Journal of biological chemistry 278 : 39839–39850.
28. DeRocherA, GilbertB, FeaginJE, ParsonsM (2005) Dissection of brefeldin A-sensitive and -insensitive steps in apicoplast protein targeting. J Cell Sci 118 : 565–574.
29. StriepenB, CrawfordMJ, ShawMK, TilneyLG, SeeberF, et al. (2000) The plastid of Toxoplasma gondii is divided by association with the centrosomes. The Journal of cell biology 151 : 1423–1434.
30. DonaldRG, RoosDS (1995) Insertional mutagenesis and marker rescue in a protozoan parasite: cloning of the uracil phosphoribosyltransferase locus from Toxoplasma gondii. Proceedings of the National Academy of Sciences of the United States of America 92 : 5749–5753.
31. CookWJ, JeffreyLC, XuY, ChauV (1993) Tertiary structures of class I ubiquitin-conjugating enzymes are highly conserved: crystal structure of yeast Ubc4. Biochemistry 32 : 13809–13817.
32. HaasAL, SiepmannTJ (1997) Pathways of ubiquitin conjugation. FASEB journal : official publication of the Federation of American Societies for Experimental Biology 11 : 1257–1268.
33. CookBW, ShawGS (2012) Architecture of the catalytic HPN motif is conserved in all E2 conjugating enzymes. The Biochemical journal 445 : 167–174.
34. WuPY, HanlonM, EddinsM, TsuiC, RogersRS, et al. (2003) A conserved catalytic residue in the ubiquitin-conjugating enzyme family. The EMBO journal 22 : 5241–5250.
35. GallagherJR, MatthewsKA, PriggeST (2011) Plasmodium falciparum apicoplast transit peptides are unstructured in vitro and during apicoplast import. Traffic 12 : 1124–1138.
36. LingQ, HuangW, BaldwinA, JarvisP (2012) Chloroplast biogenesis is regulated by direct action of the ubiquitin-proteasome system. Science 338 : 655–659.
37. KarnatakiA, DeRocherAE, FeaginJE, ParsonsM (2009) Sequential processing of the Toxoplasma apicoplast membrane protein FtsH1 in topologically distinct domains during intracellular trafficking. Mol Biochem Parasitol 166 : 126–133.
38. SchulzeA, StanderaS, BuergerE, KikkertM, van VoordenS, et al. (2005) The ubiquitin-domain protein HERP forms a complex with components of the endoplasmic reticulum associated degradation pathway. Journal of molecular biology 354 : 1021–1027.
39. KnyM, StanderaS, Hartmann-PetersenR, KloetzelPM, SeegerM (2011) Herp regulates Hrd1-mediated ubiquitylation in a ubiquitin-like domain-dependent manner. The Journal of biological chemistry 286 : 5151–5156.
40. HoellerD, DikicI (2009) Targeting the ubiquitin system in cancer therapy. Nature 458 : 438–444.
41. Le RochKG, ZhouY, BlairPL, GraingerM, MochJK, et al. (2003) Discovery of gene function by expression profiling of the malaria parasite life cycle. Science 301 : 1503–1508.
42. MazumdarJ, EHW, MasekK, CAH, StriepenB (2006) Apicoplast fatty acid synthesis is essential for organelle biogenesis and parasite survival in Toxoplasma gondii. Proceedings of the National Academy of Sciences of the United States of America 103 : 13192–13197.
43. CrawfordMJ, Thomsen-ZiegerN, RayM, SchachtnerJ, RoosDS, et al. (2006) Toxoplasma gondii scavenges host-derived lipoic acid despite its de novo synthesis in the apicoplast. The EMBO journal 25 : 3214–3222.
Štítky
Hygiena a epidemiológia Infekčné lekárstvo Laboratórium
Článok vyšiel v časopisePLOS Pathogens
Najčítanejšie tento týždeň
2013 Číslo 6- Parazitičtí červi v terapii Crohnovy choroby a dalších zánětlivých autoimunitních onemocnění
- Očkování proti virové hemoragické horečce Ebola experimentální vakcínou rVSVDG-ZEBOV-GP
- Koronavirus hýbe světem: Víte jak se chránit a jak postupovat v případě podezření?
-
Všetky články tohto čísla
- Asthma and the Diversity of Fungal Spores in Air
- The Spatial Organization of DNA Virus Genomes in the Nucleus
- Novel Polyomaviruses of Nonhuman Primates: Genetic and Serological Predictors for the Existence of Multiple Unknown Polyomaviruses within the Human Population
- Introducing the Outbreak Threshold in Epidemiology
- An Apicoplast Localized Ubiquitylation System Is Required for the Import of Nuclear-encoded Plastid Proteins
- Streptolysin O and its Co-Toxin NAD-glycohydrolase Protect Group A from Xenophagic Killing
- A Type IV Pilus Mediates DNA Binding during Natural Transformation in
- Location of the CD8 T Cell Epitope within the Antigenic Precursor Determines Immunogenicity and Protection against the Parasite
- Dietary Cholesterol Modulates Pathogen Blocking by
- Extreme Genetic Fragility of the HIV-1 Capsid
- Novel Immunomodulators from Hard Ticks Selectively Reprogramme Human Dendritic Cell Responses
- Extramedullary Myelopoiesis in Malaria Depends on Mobilization of Myeloid-Restricted Progenitors by IFN-γ Induced Chemokines
- Developing Models of Disease Transmission: Insights from Ecological Studies of Insects and Their Baculoviruses
- Cryotomography of Budding Influenza A Virus Reveals Filaments with Diverse Morphologies that Mostly Do Not Bear a Genome at Their Distal End
- PLOS Pathogens
- Archív čísel
- Aktuálne číslo
- Informácie o časopise
Najčítanejšie v tomto čísle- Asthma and the Diversity of Fungal Spores in Air
- Streptolysin O and its Co-Toxin NAD-glycohydrolase Protect Group A from Xenophagic Killing
- Cryotomography of Budding Influenza A Virus Reveals Filaments with Diverse Morphologies that Mostly Do Not Bear a Genome at Their Distal End
- A Type IV Pilus Mediates DNA Binding during Natural Transformation in
Prihlásenie#ADS_BOTTOM_SCRIPTS#Zabudnuté hesloZadajte e-mailovú adresu, s ktorou ste vytvárali účet. Budú Vám na ňu zasielané informácie k nastaveniu nového hesla.
- Časopisy



